Distinct spatiotemporal dynamics of CD8+ T cell-derived cytokines in the tumor microenvironment
- PMID: 38194914
- PMCID: PMC10783802
- DOI: 10.1016/j.ccell.2023.12.010
Distinct spatiotemporal dynamics of CD8+ T cell-derived cytokines in the tumor microenvironment
Abstract
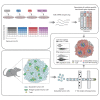
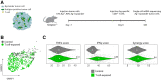
Cells in the tumor microenvironment (TME) influence each other through secretion and sensing of soluble mediators, such as cytokines and chemokines. While signaling of interferon γ (IFNγ) and tumor necrosis factor α (TNFα) is integral to anti-tumor immune responses, our understanding of the spatiotemporal behavior of these cytokines is limited. Here, we describe a single cell transcriptome-based approach to infer which signal(s) an individual cell has received. We demonstrate that, contrary to expectations, CD8+ T cell-derived IFNγ is the dominant modifier of the TME relative to TNFα. Furthermore, we demonstrate that cell pools that show abundant IFNγ sensing are characterized by decreased expression of transforming growth factor β (TGFβ)-induced genes, consistent with IFNγ-mediated TME remodeling. Collectively, these data provide evidence that CD8+ T cell-secreted cytokines should be categorized into local and global tissue modifiers, and describe a broadly applicable approach to dissect cytokine and chemokine modulation of the TME.
Keywords: IFN-gamma; T cell; TNF-alpha; cytokine; single cell RNA-seq; tumor microenvironment.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.F.A.W. received project funding for unrelated work from Bristoll-Myers-Squibb. T.N.S. is advisor for Allogene Therapeutics, Asher Bio, Merus, Neogene Therapeutics, and Scenic Biotech; is a stockholder in Allogene Therapeutics, Asher Bio, Cell Control, Celsius, Merus, and Scenic Biotech; and is venture partner at Third Rock Ventures, all outside of the current work.
Figures



References
-
- Giladi A., Cohen M., Medaglia C., Baran Y., Li B., Zada M., Bost P., Blecher-Gonen R., Salame T.M., Mayer J.U., et al. Dissecting cellular crosstalk by sequencing physically interacting cells. Nat. Biotechnol. 2020;38:629–637. - PubMed
-
- Rosenberg S.A., Yang J.C., Sherry R.M., Kammula U.S., Hughes M.S., Phan G.Q., Citrin D.E., Restifo N.P., Robbins P.F., Wunderlich J.R., et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 2011;17:4550–4557. - PMC - PubMed
-
- Dudley M.E., Gross C.A., Somerville R.P.T., Hong Y., Schaub N.P., Rosati S.F., White D.E., Nathan D., Restifo N.P., Steinberg S.M., et al. Randomized selection design trial evaluating CD8+-enriched versus unselected tumor-infiltrating lymphocytes for adoptive cell therapy for patients with melanoma. J. Clin. Oncol. 2013;31:2152–2159. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

