Multi-omics of the gut microbial ecosystem in patients with microsatellite-instability-high gastrointestinal cancer resistant to immunotherapy
- PMID: 38194971
- PMCID: PMC10829783
- DOI: 10.1016/j.xcrm.2023.101355
Multi-omics of the gut microbial ecosystem in patients with microsatellite-instability-high gastrointestinal cancer resistant to immunotherapy
Abstract
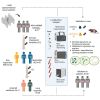
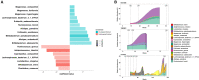
Despite the encouraging efficacy of anti-PD-1/PD-L1 immunotherapy in microsatellite-instability-high/deficient mismatch repair (MSI-H/dMMR) advanced gastrointestinal cancer, many patients exhibit primary or acquired resistance. Using multi-omics approaches, we interrogate gut microbiome, blood metabolome, and cytokines/chemokines of patients with MSI-H/dMMR gastrointestinal cancer (N = 77) at baseline and during the treatment. We identify a number of microbes (e.g., Porphyromonadaceae) and metabolites (e.g., arginine) highly associated with primary resistance to immunotherapy. An independent validation cohort (N = 39) and mouse model are used to further confirm our findings. A predictive machine learning model for primary resistance is also built and achieves an accuracy of 0.79 on the external validation set. Furthermore, several microbes are pinpointed that gradually changed during the process of acquired resistance. In summary, our study demonstrates the essential role of gut microbiome in drug resistance, and this can be utilized as a preventative diagnosis tool and therapeutic target in the future.
Keywords: MSI-H/dMMR; gastrointestinal cancer; gut microbiome; immunotherapy; microsatellite instability-high/deficient mismatch repair.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Poynter J.N., Siegmund K.D., Weisenberger D.J., Long T.I., Thibodeau S.N., Lindor N., Young J., Jenkins M.A., Hopper J.L., Baron J.A., et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol. Biomarkers Prev. 2008;17:3208–3215. - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials

