Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE
- PMID: 38195637
- PMCID: PMC10776854
- DOI: 10.1038/s41467-023-44670-5
Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE
Abstract
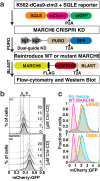
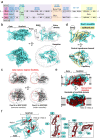
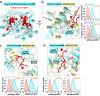
Transmembrane E3 ligases play crucial roles in homeostasis. Much protein and organelle quality control, and metabolic regulation, are determined by ER-resident MARCH6 E3 ligases, including Doa10 in yeast. Here, we present Doa10/MARCH6 structural analysis by cryo-EM and AlphaFold predictions, and a structure-based mutagenesis campaign. The majority of Doa10/MARCH6 adopts a unique circular structure within the membrane. This channel is established by a lipid-binding scaffold, and gated by a flexible helical bundle. The ubiquitylation active site is positioned over the channel by connections between the cytosolic E3 ligase RING domain and the membrane-spanning scaffold and gate. Here, by assaying 95 MARCH6 variants for effects on stability of the well-characterized substrate SQLE, which regulates cholesterol levels, we reveal crucial roles of the gated channel and RING domain consistent with AlphaFold-models of substrate-engaged and ubiquitylation complexes. SQLE degradation further depends on connections between the channel and RING domain, and lipid binding sites, revealing how interconnected Doa10/MARCH6 elements could orchestrate metabolic signals, substrate binding, and E3 ligase activity.
© 2024. The Author(s).
Conflict of interest statement
B.A.S. is adjunct faculty at St. Jude Children’s Research Hospital, Memphis, TN, USA, is on the Scientific Advisory Boards of BioTheryX and Proxygen, and is co-inventor of intellectual property related to DCN1 inhibitors (unrelated to this work) licensed to Cinsano. M.A.S. is co-founder and shareholder of Linkster Therapeutics AG. The remaining authors declare no competing interest.
Figures




References
MeSH terms
Substances
Grants and funding
- Schulman Department/Max-Planck-Gesellschaft (Max Planck Society)
- Schulman Department/Max-Planck-Gesellschaft (Max Planck Society)
- Schulman Department/Max-Planck-Gesellschaft (Max Planck Society)
- Murray Group/Max-Planck-Gesellschaft (Max Planck Society)
- UPSmeetMet, 101098161/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- ERC AdvG, UPSmeetMet, 101098161/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- Redox in macrophages, 101062335/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- DFG BR 6742/1-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources

