Upregulated pexophagy limits the capacity of selective autophagy
- PMID: 38195640
- PMCID: PMC10776696
- DOI: 10.1038/s41467-023-44005-4
Upregulated pexophagy limits the capacity of selective autophagy
Abstract
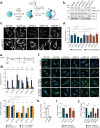
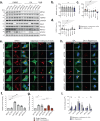
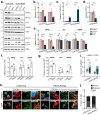
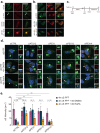
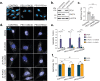
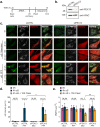
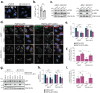
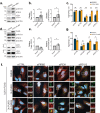
Selective autophagy is an essential process to maintain cellular homeostasis through the constant recycling of damaged or superfluous components. Over a dozen selective autophagy pathways mediate the degradation of diverse cellular substrates, but whether these pathways can influence one another remains unknown. We address this question using pexophagy, the autophagic degradation of peroxisomes, as a model. We show in cells that upregulated pexophagy impairs the selective autophagy of both mitochondria and protein aggregates by exhausting the autophagy initiation factor, ULK1. We confirm this finding in cell models of the pexophagy-mediated form of Zellweger Spectrum Disorder, a disease characterized by peroxisome dysfunction. Further, we extend the generalizability of limited selective autophagy by determining that increased protein aggregate degradation reciprocally reduces pexophagy using cell models of Parkinson's Disease and Huntington's Disease. Our findings suggest that the degradative capacity of selective autophagy can become limited by an increase in one substrate.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

