Breaking NGF-TrkA immunosuppression in melanoma sensitizes immunotherapy for durable memory T cell protection
- PMID: 38195702
- PMCID: PMC11377935
- DOI: 10.1038/s41590-023-01723-7
Breaking NGF-TrkA immunosuppression in melanoma sensitizes immunotherapy for durable memory T cell protection
Abstract
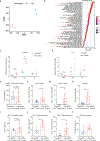
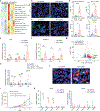
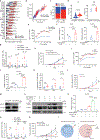
Melanoma cells, deriving from neuroectodermal melanocytes, may exploit the nervous system's immune privilege for growth. Here we show that nerve growth factor (NGF) has both melanoma cell intrinsic and extrinsic immunosuppressive functions. Autocrine NGF engages tropomyosin receptor kinase A (TrkA) on melanoma cells to desensitize interferon γ signaling, leading to T and natural killer cell exclusion. In effector T cells that upregulate surface TrkA expression upon T cell receptor activation, paracrine NGF dampens T cell receptor signaling and effector function. Inhibiting NGF, either through genetic modification or with the tropomyosin receptor kinase inhibitor larotrectinib, renders melanomas susceptible to immune checkpoint blockade therapy and fosters long-term immunity by activating memory T cells with low affinity. These results identify the NGF-TrkA axis as an important suppressor of anti-tumor immunity and suggest larotrectinib might be repurposed for immune sensitization. Moreover, by enlisting low-affinity T cells, anti-NGF reduces acquired resistance to immune checkpoint blockade and prevents melanoma recurrence.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
Q.J.L was a scientific co-founder and is a shareholder of TCRCure Biopharma and Hervor Therapeutics. The other authors declare no conflict of interests.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

