Cerebrospinal fluid proteomics in patients with Alzheimer's disease reveals five molecular subtypes with distinct genetic risk profiles
- PMID: 38195725
- PMCID: PMC10798889
- DOI: 10.1038/s43587-023-00550-7
Cerebrospinal fluid proteomics in patients with Alzheimer's disease reveals five molecular subtypes with distinct genetic risk profiles
Abstract
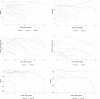
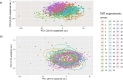
Alzheimer's disease (AD) is heterogenous at the molecular level. Understanding this heterogeneity is critical for AD drug development. Here we define AD molecular subtypes using mass spectrometry proteomics in cerebrospinal fluid, based on 1,058 proteins, with different levels in individuals with AD (n = 419) compared to controls (n = 187). These AD subtypes had alterations in protein levels that were associated with distinct molecular processes: subtype 1 was characterized by proteins related to neuronal hyperplasticity; subtype 2 by innate immune activation; subtype 3 by RNA dysregulation; subtype 4 by choroid plexus dysfunction; and subtype 5 by blood-brain barrier impairment. Each subtype was related to specific AD genetic risk variants, for example, subtype 1 was enriched with TREM2 R47H. Subtypes also differed in clinical outcomes, survival times and anatomical patterns of brain atrophy. These results indicate molecular heterogeneity in AD and highlight the need for personalized medicine.
© 2024. The Author(s).
Conflict of interest statement
P.J.V. and B.M.T. are coinventors on a patent of CSF proteomic subtypes (published under patent no. US2022196683A1, owner VUmc Foundation). E.V. is cofounder of ADx NeuroSciences, while J.G. is an employee of ADx NeuroSciences. F.B. is on the steering committee or data safety monitoring board for Biogen, Merck, ATRI/ACTC and Prothena. He is a consultant for Roche, Celltrion, Rewind Therapeutics, Merck, IXICO, Jansen and Combinostics. He has research agreements with Merck, Biogen, GE Healthcare and Roche, and he is cofounder and shareholder of Queen Square Analytics LTD. L.V. received consulting fees from Roche and Olink, all paid to Amsterdam UMC. C.E.T. performed contract research for ADx NeuroSciences, AC-Immune, Aribio, Axon Neurosciences, Beckman-Coulter, BioConnect, Bioorchestra, Brainstorm Therapeutics, Celgene, Cognition Therapeutics, EIP Pharma, Eisai, Eli Lilly, Fujirebio, Grifols, Instant Nano Biosensors, Merck, Novo Nordisk, Olink, PeopleBio, Quanterix, Roche, Siemens, Toyama and Vivoryon. She is editor of
Figures






References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

