IgM N-glycosylation correlates with COVID-19 severity and rate of complement deposition
- PMID: 38195739
- PMCID: PMC10776791
- DOI: 10.1038/s41467-023-44211-0
IgM N-glycosylation correlates with COVID-19 severity and rate of complement deposition
Abstract
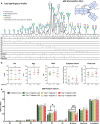
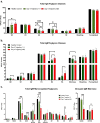
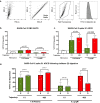
The glycosylation of IgG plays a critical role during human severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, activating immune cells and inducing cytokine production. However, the role of IgM N-glycosylation has not been studied during human acute viral infection. The analysis of IgM N-glycosylation from healthy controls and hospitalized coronavirus disease 2019 (COVID-19) patients reveals increased high-mannose and sialylation that correlates with COVID-19 severity. These trends are confirmed within SARS-CoV-2-specific immunoglobulin N-glycan profiles. Moreover, the degree of total IgM mannosylation and sialylation correlate significantly with markers of disease severity. We link the changes of IgM N-glycosylation with the expression of Golgi glycosyltransferases. Lastly, we observe antigen-specific IgM antibody-dependent complement deposition is elevated in severe COVID-19 patients and modulated by exoglycosidase digestion. Taken together, this work links the IgM N-glycosylation with COVID-19 severity and highlights the need to understand IgM glycosylation and downstream immune function during human disease.
© 2024. The Author(s).
Conflict of interest statement
The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list Florian Krammer (F.K.) as co-inventor. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. F.K. has consulted for Merck, Seqirus, Curevac and Pfizer, and is currently consulting for GSK, Gritstone, 3rd Rock Ventures and Avimex and he is a co-founder and scientific advisory board member of CastleVax. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2 and Dynavax on influenza virus vaccines. All other authors have no competing interests to declare.
Figures



Update of
-
IgM N-glycosylation correlates with COVID-19 severity and rate of complement deposition.Res Sq [Preprint]. 2023 Jun 2:rs.3.rs-2939468. doi: 10.21203/rs.3.rs-2939468/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Jan 9;15(1):404. doi: 10.1038/s41467-023-44211-0. PMID: 37398192 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- U19 AI090023/AI/NIAID NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- S10 OD026880/OD/NIH HHS/United States
- U01 HL163124/HL/NHLBI NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
- U19 AI167891/AI/NIAID NIH HHS/United States
- U19 AI167903/AI/NIAID NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- I01 BX005023/BX/BLRD VA/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- R01 AI132774/AI/NIAID NIH HHS/United States
- P01 AI153559/AI/NIAID NIH HHS/United States
- S10 OD030463/OD/NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

