This is a preprint.
Rare variant analyses validate known ALS genes in a multi-ethnic population and identifies ANTXR2 as a candidate in PLS
- PMID: 38196621
- PMCID: PMC10775375
- DOI: 10.21203/rs.3.rs-3721598/v1
Rare variant analyses validate known ALS genes in a multi-ethnic population and identifies ANTXR2 as a candidate in PLS
Update in
-
Rare variant analyses validate known ALS genes in a multi-ethnic population and identifies ANTXR2 as a candidate in PLS.BMC Genomics. 2024 Jun 29;25(1):651. doi: 10.1186/s12864-024-10538-1. BMC Genomics. 2024. PMID: 38951798 Free PMC article.
Abstract
Background: Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease affecting over 30,000 people in the United States. It is characterized by the progressive decline of the nervous system that leads to the weakening of muscles which impacts physical function. Approximately, 15% of individuals diagnosed with ALS have a known genetic variant that contributes to their disease. As therapies that slow or prevent symptoms, such as antisense oligonucleotides, continue to develop, it is important to discover novel genes that could be targets for treatment. Additionally, as cohorts continue to grow, performing analyses in ALS subtypes, such as primary lateral sclerosis (PLS), becomes possible due to an increase in power. These analyses could highlight novel pathways in disease manifestation.
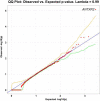
Methods: Building on our previous discoveries using rare variant association analyses, we conducted rare variant burden testing on a substantially larger cohort of 6,970 ALS patients from a large multi-ethnic cohort as well as 166 PLS patients, and 22,524 controls. We used intolerant domain percentiles based on sub-region Residual Variation Intolerance Score (subRVIS) that have been described previously in conjunction with gene based collapsing approaches to conduct burden testing to identify genes that associate with ALS and PLS.
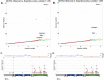
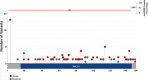
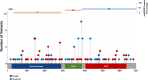
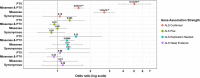
Results: A gene based collapsing model showed significant associations with SOD1, TARDBP, and TBK1 (OR=19.18, p = 3.67 × 10-39; OR=4.73, p = 2 × 10-10; OR=2.3, p = 7.49 × 10-9, respectively). These genes have been previously associated with ALS. Additionally, a significant novel control enriched gene, ALKBH3 (p = 4.88 × 10-7), was protective for ALS in this model. An intolerant domain based collapsing model showed a significant improvement in identifying regions in TARDBP that associated with ALS (OR=10.08, p = 3.62 × 10-16). Our PLS protein truncating variant collapsing analysis demonstrated significant case enrichment in ANTXR2 (p=8.38 × 10-6).
Conclusions: In a large multi-ethnic cohort of 6,970 ALS patients, rare variant burden testing validated known ALS genes and identified a novel potentially protective gene, ALKBH3. A first-ever analysis in 166 patients with PLS found a candidate association with loss-of-function mutations in ANTXR2.
Keywords: ALS; Amyotrophic lateral sclerosis; Burden testing; PLS; Rare-variant analyses; peripheral lateral sclerosis.
Conflict of interest statement
COMPETING INTERESTS The authors declare that they have no competing interests Additional Declarations: No competing interests reported.
Figures
Similar articles
-
Rare variant analyses validate known ALS genes in a multi-ethnic population and identifies ANTXR2 as a candidate in PLS.BMC Genomics. 2024 Jun 29;25(1):651. doi: 10.1186/s12864-024-10538-1. BMC Genomics. 2024. PMID: 38951798 Free PMC article.
-
Rare variant analyses validate known ALS genes in a multi-ethnic population and identifies ANTXR2 as a candidate in PLS.medRxiv [Preprint]. 2023 Oct 23:2023.09.30.23296353. doi: 10.1101/2023.09.30.23296353. medRxiv. 2023. Update in: BMC Genomics. 2024 Jun 29;25(1):651. doi: 10.1186/s12864-024-10538-1. PMID: 37873269 Free PMC article. Updated. Preprint.
-
Decoding genetic and pathophysiological mechanisms in amyotrophic lateral sclerosis and primary lateral sclerosis: A comparative study of differentially expressed genes and implicated pathways in motor neuron disorders.Adv Protein Chem Struct Biol. 2024;141:177-201. doi: 10.1016/bs.apcsb.2023.12.008. Epub 2024 Jun 25. Adv Protein Chem Struct Biol. 2024. PMID: 38960473
-
Studies of Genetic and Proteomic Risk Factors of Amyotrophic Lateral Sclerosis Inspire Biomarker Development and Gene Therapy.Cells. 2023 Jul 27;12(15):1948. doi: 10.3390/cells12151948. Cells. 2023. PMID: 37566027 Free PMC article. Review.
-
Familial motor neuron disease: co-occurrence of PLS and ALS (-FTD).Amyotroph Lateral Scler Frontotemporal Degener. 2024 Feb;25(1-2):53-60. doi: 10.1080/21678421.2023.2255621. Epub 2023 Sep 7. Amyotroph Lateral Scler Frontotemporal Degener. 2024. PMID: 37679883 Review.
References
Publication types
Grants and funding
- K01 MH098126/MH/NIMH NIH HHS/United States
- MC_PC_15080/MRC_/Medical Research Council/United Kingdom
- RC2 MH089915/MH/NIMH NIH HHS/United States
- P01 HD080642/HD/NICHD NIH HHS/United States
- R01 MH097971/MH/NIMH NIH HHS/United States
- RC2 NS070344/NS/NINDS NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- UL1 TR001873/TR/NCATS NIH HHS/United States
- UM1 AI100645/AI/NIAID NIH HHS/United States
- P30 AG028377/AG/NIA NIH HHS/United States
- U54 NS078059/NS/NINDS NIH HHS/United States
- P01 AG007232/AG/NIA NIH HHS/United States
- RF1 AG054023/AG/NIA NIH HHS/United States
- R01 HD048805/HD/NICHD NIH HHS/United States
- U01 HG007672/HG/NHGRI NIH HHS/United States
- U01 NS053998/NS/NINDS NIH HHS/United States
- U01 NS077303/NS/NINDS NIH HHS/United States
- U19 AI067854/AI/NIAID NIH HHS/United States
- R01 AG037212/AG/NIA NIH HHS/United States
- R01 ES016348/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

