This is a preprint.
Genome-wide QTL mapping across three tissues highlights several Alzheimer's and Parkinson's disease loci potentially acting via DNA methylation
- PMID: 38196633
- PMCID: PMC10775408
- DOI: 10.1101/2023.12.22.23300365
Genome-wide QTL mapping across three tissues highlights several Alzheimer's and Parkinson's disease loci potentially acting via DNA methylation
Abstract
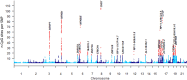
DNA methylation (DNAm) is an epigenetic mark with essential roles in disease development and predisposition. Here, we created genome-wide maps of methylation quantitative trait loci (meQTL) in three peripheral tissues and used Mendelian randomization (MR) analyses to assess the potential causal relationships between DNAm and risk for two common neurodegenerative disorders, i.e. Alzheimer's disease (AD) and Parkinson's disease (PD). Genome-wide single nucleotide polymorphism (SNP; ~5.5M sites) and DNAm (~850K CpG sites) data were generated from whole blood (n=1,058), buccal (n=1,527) and saliva (n=837) specimens. We identified between 11 and 15 million genome-wide significant (p<10-14) SNP-CpG associations in each tissue. Combining these meQTL GWAS results with recent AD/PD GWAS summary statistics by MR strongly suggests that the previously described associations between PSMC3, PICALM, and TSPAN14 and AD may be founded on differential DNAm in or near these genes. In addition, there is strong, albeit less unequivocal, support for causal links between DNAm at PRDM7 in AD as well as at KANSL1/MAPT in AD and PD. Our study adds valuable insights on AD/PD pathogenesis by combining two high-resolution "omics" domains, and the meQTL data shared along with this publication will allow like-minded analyses in other diseases.
Keywords: Alzheimer’s disease (AD); Genome-wide association study (GWAS); Mendelian randomization (MR); Parkinson’s disease (PD); colocalization; methylation quantitative trait locus (meQTL) analysis.
Conflict of interest statement
Competing interests D.B.F. serves on the scientific advisory board of Linus Health. A.P.L. serves on the scientific advisory boards for Neuroelectrics, Magstim Inc., TetraNeuron, Skin2Neuron, MedRhythms, and Hearts Radiant. He is co-founder of TI solutions and co-founder and chief medical officer of Linus Health. Furthermore, A.P.L. is listed as an inventor on several issued and pending patents on the real-time integration of transcranial magnetic stimulation with electroencephalography and magnetic resonance imaging, and applications of noninvasive brain stimulation in various neurological disorders; as well as digital biomarkers of cognition and digital assessments for early diagnosis of dementia. The remaining authors declare no competing interests.
Figures







References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
