Large scale sequence-based screen for recessive variants allows for identification and monitoring of rare deleterious variants in pigs
- PMID: 38198533
- PMCID: PMC10805306
- DOI: 10.1371/journal.pgen.1011034
Large scale sequence-based screen for recessive variants allows for identification and monitoring of rare deleterious variants in pigs
Abstract
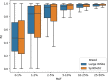
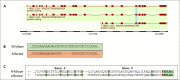
Most deleterious variants are recessive and segregate at relatively low frequency. Therefore, high sample sizes are required to identify these variants. In this study we report a large-scale sequence based genome-wide association study (GWAS) in pigs, with a total of 120,000 Large White and 80,000 Synthetic breed animals imputed to sequence using a reference population of approximately 1,100 whole genome sequenced pigs. We imputed over 20 million variants with high accuracies (R2>0.9) even for low frequency variants (1-5% minor allele frequency). This sequence-based analysis revealed a total of 14 additive and 9 non-additive significant quantitative trait loci (QTLs) for growth rate and backfat thickness. With the non-additive (recessive) model, we identified a deleterious missense SNP in the CDHR2 gene reducing growth rate and backfat in homozygous Large White animals. For the Synthetic breed, we revealed a QTL on chromosome 15 with a frameshift variant in the OBSL1 gene. This QTL has a major impact on both growth rate and backfat, resembling human 3M-syndrome 2 which is related to the same gene. With the additive model, we confirmed known QTLs on chromosomes 1 and 5 for both breeds, including variants in the MC4R and CCND2 genes. On chromosome 1, we disentangled a complex QTL region with multiple variants affecting both traits, harboring 4 independent QTLs in the span of 5 Mb. Together we present a large scale sequence-based association study that provides a key resource to scan for novel variants at high resolution for breeding and to further reduce the frequency of deleterious alleles at an early stage in the breeding program.
Copyright: © 2024 Boshove et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
All authors declare that the results are presented in full and present no conflict of interest.
Figures



Similar articles
-
Comparison of two multi-trait association testing methods and sequence-based fine mapping of six additive QTL in Swiss Large White pigs.BMC Genomics. 2023 Apr 10;24(1):192. doi: 10.1186/s12864-023-09295-4. BMC Genomics. 2023. PMID: 37038103 Free PMC article.
-
Large-scale association study on daily weight gain in pigs reveals overlap of genetic factors for growth in humans.BMC Genomics. 2022 Feb 15;23(1):133. doi: 10.1186/s12864-022-08373-3. BMC Genomics. 2022. PMID: 35168569 Free PMC article.
-
An association study using imputed whole-genome sequence data identifies novel significant loci for growth-related traits in a Duroc × Erhualian F2 population.J Anim Breed Genet. 2019 May;136(3):217-228. doi: 10.1111/jbg.12389. Epub 2019 Mar 14. J Anim Breed Genet. 2019. PMID: 30869175
-
Meta-analysis of sequence-based association studies across three cattle breeds reveals 25 QTL for fat and protein percentages in milk at nucleotide resolution.BMC Genomics. 2017 Nov 9;18(1):853. doi: 10.1186/s12864-017-4263-8. BMC Genomics. 2017. PMID: 29121857 Free PMC article.
-
A multi-trait meta-analysis with imputed sequence variants reveals twelve QTL for mammary gland morphology in Fleckvieh cattle.Genet Sel Evol. 2016 Feb 16;48:14. doi: 10.1186/s12711-016-0190-4. Genet Sel Evol. 2016. PMID: 26883850 Free PMC article.
Cited by
-
Genome-Wide Association Study for Individual Primal Cut Quality Traits in Canadian Commercial Crossbred Pigs.Animals (Basel). 2025 Jun 13;15(12):1754. doi: 10.3390/ani15121754. Animals (Basel). 2025. PMID: 40564306 Free PMC article.
-
Genome-Wide Association Study for Belly Traits in Canadian Commercial Crossbred Pigs.Animals (Basel). 2025 Apr 29;15(9):1254. doi: 10.3390/ani15091254. Animals (Basel). 2025. PMID: 40362070 Free PMC article.
References
-
- Sariya S, Lee JH, Mayeux R, Vardarajan BN, Reyes-Dumeyer D, Manly JJ, et al.. Rare Variants Imputation in Admixed Populations: Comparison Across Reference Panels and Bioinformatics Tools. Front Genet. 2019;10. Available: https://www.frontiersin.org/articles/10.3389/fgene.2019.00239 - DOI - PMC - PubMed
-
- Qanbari S. On the Extent of Linkage Disequilibrium in the Genome of Farm Animals. Front Genet. 2020;10. Available: https://www.frontiersin.org/articles/10.3389/fgene.2019.01304 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

