RREB1 regulates neuronal proteostasis and the microtubule network
- PMID: 38198538
- PMCID: PMC10780896
- DOI: 10.1126/sciadv.adh3929
RREB1 regulates neuronal proteostasis and the microtubule network
Abstract
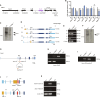
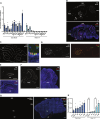
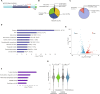
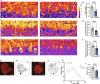
Transcription factors play vital roles in neuron development; however, little is known about the role of these proteins in maintaining neuronal homeostasis. Here, we show that the transcription factor RREB1 (Ras-responsive element-binding protein 1) is essential for neuron survival in the mammalian brain. A spontaneous mouse mutation causing loss of a nervous system-enriched Rreb1 transcript is associated with progressive loss of cerebellar Purkinje cells and ataxia. Analysis of chromatin immunoprecipitation and sequencing, along with RNA sequencing data revealed dysregulation of RREB1 targets associated with the microtubule cytoskeleton. In agreement with the known role of microtubules in dendritic development, dendritic complexity was disrupted in Rreb1-deficient neurons. Analysis of sequencing data also suggested that RREB1 plays a role in the endomembrane system. Mutant Purkinje cells had fewer numbers of autophagosomes and lysosomes and contained P62- and ubiquitin-positive inclusions. Together, these studies demonstrate that RREB1 functions to maintain the microtubule network and proteostasis in mammalian neurons.
Figures

References
-
- Gu Z., Cao H., Zuo C., Huang Y., Miao J., Song Y., Yang Y., Zhu L., Wang F., TFEB in Alzheimer’s disease: From molecular mechanisms to therapeutic implications. Neurobiol. Dis. 173, 105855 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

