LncEDCH1 g.1703613 T>C regulates chicken carcass traits by targeting miR-196-2-3p
- PMID: 38198912
- PMCID: PMC10825527
- DOI: 10.1016/j.psj.2023.103412
LncEDCH1 g.1703613 T>C regulates chicken carcass traits by targeting miR-196-2-3p
Abstract
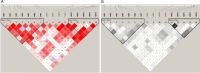
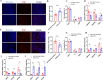
Single nucleotide polymorphisms (SNPs) are valuable genetic markers that can provide insights into the genetic diversity and variation within chicken populations. In poultry breeding, SNP analysis is widely utilized to accelerate the selection of desirable traits, improving the efficiency and effectiveness of chicken breeding programs. In our previous research, we identified an association between LncEDCH1 and muscle development. To further investigate its specific mechanism, we conducted SNP detection and performed genotyping, linkage disequilibrium, and haplotype analysis. Our research findings indicate that 16 SNPs in the LncEDCH1. Among these SNPs, g.1703497 C>T and g.1704262 C>T were significantly associated with breast muscle weight percentage, g.1703497 C>T and g.1703613 T>C were significantly associated with leg weight percentage, and g.1703497 C>T, g.1703589 T>C, g.1703613 T>C, g.1703636 C>A, g.1703768 T>C, g.1704079 C>T, g.1704250 T>C, g.1704253 G>A were significantly associated with skin yellowness. Two haplotype blocks composed of 6 SNPs that were significantly associated with wing skin yellowness, breast skin yellowness, full-bore weight, and carcass weight percentage. Furthermore, through dual-luciferase reporter assays, biotin-coupled miRNA pull-down assays, 5-ethynyl-2'-deoxyuridine (EDU) assays, immunofluorescence, and quantitative real-time polymerase chain reaction (qPCR), it has been confirmed that miR-196-2-3p inhibits the expression of LncEDCH1 directly by binding to LncEDCH1 g.1703613T>C, thereby achieving indirect regulation of muscle development. These findings provide valuable molecular markers for chicken molecular breeding and broaden our understanding of the regulatory mechanisms.
Keywords: LncEDCH1; carcass trait; chicken; miR-196-2-3p; single nucleotide polymorphism.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Detection of CD36 gene polymorphism associated with chicken carcass traits and skin yellowness.Poult Sci. 2023 Jul;102(7):102691. doi: 10.1016/j.psj.2023.102691. Epub 2023 Apr 4. Poult Sci. 2023. PMID: 37120870 Free PMC article.
-
Detection of SNPs in the TBC1D1 gene and their association with carcass traits in chicken.Gene. 2014 Sep 1;547(2):288-94. doi: 10.1016/j.gene.2014.06.061. Epub 2014 Jun 27. Gene. 2014. PMID: 24979340
-
Identification and characterization of SREBF2 expression and its association with chicken carcass traits.Genet Mol Res. 2016 Sep 2;15(3). doi: 10.4238/gmr.15038514. Genet Mol Res. 2016. PMID: 27706686
-
Polymorphisms and expression of the chicken POU1F1 gene associated with carcass traits.Mol Biol Rep. 2012 Aug;39(8):8363-71. doi: 10.1007/s11033-012-1686-9. Epub 2012 Jun 22. Mol Biol Rep. 2012. PMID: 22722987
-
Polymorphisms in Wnt signaling pathway genes are significantly associated with chicken carcass traits.Poult Sci. 2012 Jun;91(6):1299-307. doi: 10.3382/ps.2012-02157. Poult Sci. 2012. PMID: 22582286
Cited by
-
Genetic Effects of Chicken Pre-miR-3528 SNP on Growth Performance, Meat Quality Traits, and Serum Enzyme Activities.Animals (Basel). 2025 Aug 6;15(15):2300. doi: 10.3390/ani15152300. Animals (Basel). 2025. PMID: 40805090 Free PMC article.
-
Identification of key genes associated with residual feed intake in small-sized meat ducks through integrated analysis of mRNA and miRNA transcriptomes.Poult Sci. 2025 May;104(5):105058. doi: 10.1016/j.psj.2025.105058. Epub 2025 Mar 21. Poult Sci. 2025. PMID: 40132315 Free PMC article.
References
-
- Ardlie K.G., Kruglyak L., Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 2002;3:299–309. - PubMed
-
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. - PubMed
-
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Brookes A.J. The essence of SNPs. Gene. 1999;234:177–186. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

