Methylation of elongation factor 1A by yeast Efm4 or human eEF1A-KMT2 involves a beta-hairpin recognition motif and crosstalks with phosphorylation
- PMID: 38199565
- PMCID: PMC10844748
- DOI: 10.1016/j.jbc.2024.105639
Methylation of elongation factor 1A by yeast Efm4 or human eEF1A-KMT2 involves a beta-hairpin recognition motif and crosstalks with phosphorylation
Abstract
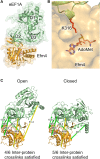
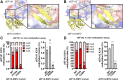
Translation elongation factor 1A (eEF1A) is an essential and highly conserved protein required for protein synthesis in eukaryotes. In both Saccharomyces cerevisiae and human, five different methyltransferases methylate specific residues on eEF1A, making eEF1A the eukaryotic protein targeted by the highest number of dedicated methyltransferases after histone H3. eEF1A methyltransferases are highly selective enzymes, only targeting eEF1A and each targeting just one or two specific residues in eEF1A. However, the mechanism of this selectivity remains poorly understood. To reveal how S. cerevisiae elongation factor methyltransferase 4 (Efm4) specifically methylates eEF1A at K316, we have used AlphaFold-Multimer modeling in combination with crosslinking mass spectrometry (XL-MS) and enzyme mutagenesis. We find that a unique beta-hairpin motif, which extends out from the core methyltransferase fold, is important for the methylation of eEF1A K316 in vitro. An alanine mutation of a single residue on this beta-hairpin, F212, significantly reduces Efm4 activity in vitro and in yeast cells. We show that the equivalent residue in human eEF1A-KMT2 (METTL10), F220, is also important for its activity towards eEF1A in vitro. We further show that the eEF1A guanine nucleotide exchange factor, eEF1Bα, inhibits Efm4 methylation of eEF1A in vitro, likely due to competitive binding. Lastly, we find that phosphorylation of eEF1A at S314 negatively crosstalks with Efm4-mediated methylation of K316. Our findings demonstrate how protein methyltransferases can be highly selective towards a single residue on a single protein in the cell.
Keywords: AlphaFold; crosslinking mass spectrometry; protein cross-linking; protein methylation; protein methyltransferase; translation elongation factor.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures






References
-
- Di Blasi R., Blyuss O., Timms J.F., Conole D., Ceroni F., Whitwell H.J. Non-histone protein methylation: biological significance and Bioengineering potential. ACS Chem. Biol. 2021;16:238–250. - PubMed
-
- Biggar K.K., Li S.S. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015;16:5–17. - PubMed
-
- Guccione E., Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019;20:642–657. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

