TRIM22 induces cellular senescence by targeting PHLPP2 in hepatocellular carcinoma
- PMID: 38199981
- PMCID: PMC10781680
- DOI: 10.1038/s41419-024-06427-w
TRIM22 induces cellular senescence by targeting PHLPP2 in hepatocellular carcinoma
Abstract
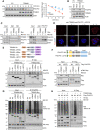
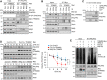
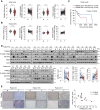
The ubiquitin-proteasome system is a vital protein degradation system that is involved in various cellular processes, such as cell cycle progression, apoptosis, and differentiation. Dysregulation of this system has been implicated in numerous diseases, including cancer, vascular disease, and neurodegenerative disorders. Induction of cellular senescence in hepatocellular carcinoma (HCC) is a potential anticancer strategy, but the precise role of the ubiquitin-proteasome system in cellular senescence remains unclear. In this study, we show that the E3 ubiquitin ligase, TRIM22, plays a critical role in the cellular senescence of HCC cells. TRIM22 expression is transcriptionally upregulated by p53 in HCC cells experiencing ionizing radiation (IR)-induced senescence. Overexpression of TRIM22 triggers cellular senescence by targeting the AKT phosphatase, PHLPP2. Mechanistically, the SPRY domain of TRIM22 directly associates with the C-terminal domain of PHLPP2, which contains phosphorylation sites that are subject to IKKβ-mediated phosphorylation. The TRIM22-mediated PHLPP2 degradation leads to activation of AKT-p53-p21 signaling, ultimately resulting in cellular senescence. In both human HCC databases and patient specimens, the levels of TRIM22 and PHLPP2 show inverse correlations at the mRNA and protein levels. Collectively, our findings reveal that TRIM22 regulates cancer cell senescence by modulating the proteasomal degradation of PHLPP2 in HCC cells, suggesting that TRIM22 could potentially serve as a therapeutic target for treating cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 2021R1A5A2031612/National Research Foundation of Korea (NRF)
- 2023R1A2C2003673/National Research Foundation of Korea (NRF)
- 2021R1A5A2031612/National Research Foundation of Korea (NRF)
- 2023R1A2C2003673/National Research Foundation of Korea (NRF)
- 2021R1A5A2031612/National Research Foundation of Korea (NRF)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

