Assembly of a unique membrane complex in type VI secretion systems of Bacteroidota
- PMID: 38200008
- PMCID: PMC10781749
- DOI: 10.1038/s41467-023-44426-1
Assembly of a unique membrane complex in type VI secretion systems of Bacteroidota
Abstract
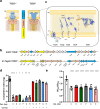
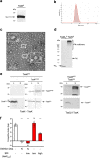
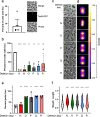
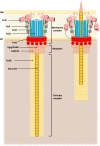
The type VI secretion system (T6SS) of Gram-negative bacteria inhibits competitor cells through contact-dependent translocation of toxic effector proteins. In Proteobacteria, the T6SS is anchored to the cell envelope through a megadalton-sized membrane complex (MC). However, the genomes of Bacteroidota with T6SSs appear to lack genes encoding homologs of canonical MC components. Here, we identify five genes in Bacteroides fragilis (tssNQOPR) that are essential for T6SS function and encode a Bacteroidota-specific MC. We purify this complex, reveal its dimensions using electron microscopy, and identify a protein-protein interaction network underlying the assembly of the MC including the stoichiometry of the five TssNQOPR components. Protein TssN mediates the connection between the Bacteroidota MC and the conserved baseplate. Although MC gene content and organization varies across the phylum Bacteroidota, no MC homologs are detected outside of T6SS loci, suggesting ancient co-option and functional convergence with the non-homologous MC of Pseudomonadota.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

