Candidate biomarkers of EV-microRNA in detecting REM sleep behavior disorder and Parkinson's disease
- PMID: 38200052
- PMCID: PMC10781790
- DOI: 10.1038/s41531-023-00628-4
Candidate biomarkers of EV-microRNA in detecting REM sleep behavior disorder and Parkinson's disease
Abstract
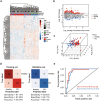
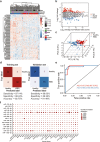
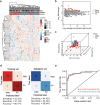
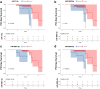
Parkinson's disease (PD) lacks reliable, non-invasive biomarker tests for early intervention and management. Thus, a minimally invasive test for the early detection and monitoring of PD and REM sleep behavior disorder (iRBD) is a highly unmet need for developing drugs and planning patient care. Extracellular vehicles (EVs) are found in a wide variety of biofluids, including plasma. EV-mediated functional transfer of microRNAs (miRNAs) may be viable candidates as biomarkers for PD and iRBD. Next-generation sequencing (NGS) of EV-derived small RNAs was performed in 60 normal controls, 56 iRBD patients and 53 PD patients to profile small non-coding RNAs (sncRNAs). Moreover, prospective follow-up was performed for these 56 iRBD patients for an average of 3.3 years. Full-scale miRNA profiles of plasma EVs were evaluated by machine-learning methods. After optimizing the library construction method for low RNA inputs (named EVsmall-seq), we built a machine learning algorithm that identified diagnostic miRNA signatures for distinguishing iRBD patients (AUC 0.969) and PD patients (AUC 0.916) from healthy individuals; and PD patients (AUC 0.929) from iRBD patients. We illustrated all the possible expression patterns across healthy-iRBD-PD hierarchy. We also showed 20 examples of miRNAs with consistently increasing or decreasing expression levels from controls to iRBD to PD. In addition, four miRNAs were found to be correlated with iRBD conversion. Distinct characteristics of the miRNA profiles among normal, iRBD and PD samples were discovered, which provides a panel of promising biomarkers for the identification of PD patients and those in the prodromal stage iRBD.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

