Breaking barriers in Candida spp. detection with Electronic Noses and artificial intelligence
- PMID: 38200060
- PMCID: PMC10781724
- DOI: 10.1038/s41598-023-50332-9
Breaking barriers in Candida spp. detection with Electronic Noses and artificial intelligence
Abstract
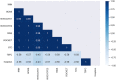
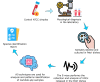
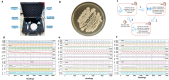
The timely and accurate diagnosis of candidemia, a severe bloodstream infection caused by Candida spp., remains challenging in clinical practice. Blood culture, the current gold standard technique, suffers from lengthy turnaround times and limited sensitivity. To address these limitations, we propose a novel approach utilizing an Electronic Nose (E-nose) combined with Time Series-based classification techniques to analyze and identify Candida spp. rapidly, using culture species of C. albicans, C.kodamaea ohmeri, C. glabrara, C. haemulonii, C. parapsilosis and C. krusei as control samples. This innovative method not only enhances diagnostic accuracy and reduces decision time for healthcare professionals in selecting appropriate treatments but also offers the potential for expanded usage and cost reduction due to the E-nose's low production costs. Our proof-of-concept experimental results, carried out with culture samples, demonstrate promising outcomes, with the Inception Time classifier achieving an impressive average accuracy of 97.46% during the test phase. This paper presents a groundbreaking advancement in the field, empowering medical practitioners with an efficient and reliable tool for early and precise identification of candidemia, ultimately leading to improved patient outcomes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

