Spatial IMA1 regulation restricts root iron acquisition on MAMP perception
- PMID: 38200311
- PMCID: PMC11181898
- DOI: 10.1038/s41586-023-06891-y
Spatial IMA1 regulation restricts root iron acquisition on MAMP perception
Abstract
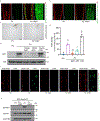
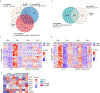
Iron is critical during host-microorganism interactions1-4. Restriction of available iron by the host during infection is an important defence strategy, described as nutritional immunity5. However, this poses a conundrum for externally facing, absorptive tissues such as the gut epithelium or the plant root epidermis that generate environments that favour iron bioavailability. For example, plant roots acquire iron mostly from the soil and, when iron deficient, increase iron availability through mechanisms that include rhizosphere acidification and secretion of iron chelators6-9. Yet, the elevated iron bioavailability would also be beneficial for the growth of bacteria that threaten plant health. Here we report that microorganism-associated molecular patterns such as flagellin lead to suppression of root iron acquisition through a localized degradation of the systemic iron-deficiency signalling peptide Iron Man 1 (IMA1) in Arabidopsis thaliana. This response is also elicited when bacteria enter root tissues, but not when they dwell on the outer root surface. IMA1 itself has a role in modulating immunity in root and shoot, affecting the levels of root colonization and the resistance to a bacterial foliar pathogen. Our findings reveal an adaptive molecular mechanism of nutritional immunity that affects iron bioavailability and uptake, as well as immune responses.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures














References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

