The CRISPR effector Cam1 mediates membrane depolarization for phage defence
- PMID: 38200316
- PMCID: PMC10808066
- DOI: 10.1038/s41586-023-06902-y
The CRISPR effector Cam1 mediates membrane depolarization for phage defence
Abstract
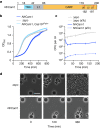
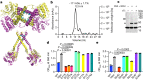
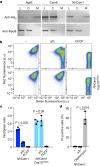
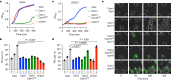
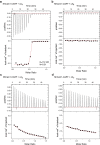
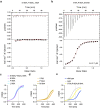
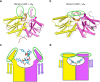
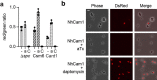
Prokaryotic type III CRISPR-Cas systems provide immunity against viruses and plasmids using CRISPR-associated Rossman fold (CARF) protein effectors1-5. Recognition of transcripts of these invaders with sequences that are complementary to CRISPR RNA guides leads to the production of cyclic oligoadenylate second messengers, which bind CARF domains and trigger the activity of an effector domain6,7. Whereas most effectors degrade host and invader nucleic acids, some are predicted to contain transmembrane helices without an enzymatic function. Whether and how these CARF-transmembrane helix fusion proteins facilitate the type III CRISPR-Cas immune response remains unknown. Here we investigate the role of cyclic oligoadenylate-activated membrane protein 1 (Cam1) during type III CRISPR immunity. Structural and biochemical analyses reveal that the CARF domains of a Cam1 dimer bind cyclic tetra-adenylate second messengers. In vivo, Cam1 localizes to the membrane, is predicted to form a tetrameric transmembrane pore, and provides defence against viral infection through the induction of membrane depolarization and growth arrest. These results reveal that CRISPR immunity does not always operate through the degradation of nucleic acids, but is instead mediated via a wider range of cellular responses.
© 2024. The Author(s).
Conflict of interest statement
L.A.M. is a co-founder of Intellia Therapeutics and Eligo Biosciences, and a scientific advisory board member of Intellia Therapeutics and Ancilia Biosciences. C.F.B., Y.Y., J.T.R., P.M. and D.J.P. declare no competing interests.
Figures





Similar articles
-
CRISPR antiphage defence mediated by the cyclic nucleotide-binding membrane protein Csx23.Nucleic Acids Res. 2024 Apr 12;52(6):2761-2775. doi: 10.1093/nar/gkae167. Nucleic Acids Res. 2024. PMID: 38471818 Free PMC article.
-
The Card1 nuclease provides defence during type III CRISPR immunity.Nature. 2021 Feb;590(7847):624-629. doi: 10.1038/s41586-021-03206-x. Epub 2021 Jan 18. Nature. 2021. PMID: 33461211 Free PMC article.
-
Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA4.J Biol Chem. 2020 Oct 30;295(44):14963-14972. doi: 10.1074/jbc.RA120.014099. Epub 2020 Aug 21. J Biol Chem. 2020. PMID: 32826317 Free PMC article.
-
The diverse arsenal of type III CRISPR-Cas-associated CARF and SAVED effectors.Biochem Soc Trans. 2022 Oct 31;50(5):1353-1364. doi: 10.1042/BST20220289. Biochem Soc Trans. 2022. PMID: 36282000 Free PMC article. Review.
-
Type III CRISPR-Cas: beyond the Cas10 effector complex.Trends Biochem Sci. 2024 Jan;49(1):28-37. doi: 10.1016/j.tibs.2023.10.006. Epub 2023 Nov 8. Trends Biochem Sci. 2024. PMID: 37949766 Free PMC article. Review.
Cited by
-
Cat1 forms filament networks to degrade NAD+ during the type III CRISPR-Cas antiviral response.Science. 2025 Jun 12;388(6752):eadv9045. doi: 10.1126/science.adv9045. Epub 2025 Jun 12. Science. 2025. PMID: 40208959 Free PMC article.
-
Mechanistic determinants and dynamics of cA6 synthesis in type III CRISPR-Cas effector complexes.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1277. doi: 10.1093/nar/gkae1277. Nucleic Acids Res. 2025. PMID: 39817514 Free PMC article.
-
Bioinformatic analysis of type III CRISPR systems reveals key properties and new effector families.Nucleic Acids Res. 2024 Jul 8;52(12):7129-7141. doi: 10.1093/nar/gkae462. Nucleic Acids Res. 2024. PMID: 38808661 Free PMC article.
-
The Cas10 nuclease activity relieves host dormancy to facilitate spacer acquisition and retention during type III-A CRISPR immunity.bioRxiv [Preprint]. 2024 Feb 12:2024.02.11.579731. doi: 10.1101/2024.02.11.579731. bioRxiv. 2024. Update in: Cell Host Microbe. 2024 Dec 11;32(12):2050-2062.e6. doi: 10.1016/j.chom.2024.11.005. PMID: 38405743 Free PMC article. Updated. Preprint.
-
CRISPR antiphage defence mediated by the cyclic nucleotide-binding membrane protein Csx23.Nucleic Acids Res. 2024 Apr 12;52(6):2761-2775. doi: 10.1093/nar/gkae167. Nucleic Acids Res. 2024. PMID: 38471818 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

