Essential genes identification model based on sequence feature map and graph convolutional neural network
- PMID: 38200437
- PMCID: PMC10777564
- DOI: 10.1186/s12864-024-09958-w
Essential genes identification model based on sequence feature map and graph convolutional neural network
Abstract
Background: Essential genes encode functions that play a vital role in the life activities of organisms, encompassing growth, development, immune system functioning, and cell structure maintenance. Conventional experimental techniques for identifying essential genes are resource-intensive and time-consuming, and the accuracy of current machine learning models needs further enhancement. Therefore, it is crucial to develop a robust computational model to accurately predict essential genes.
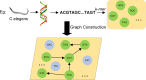
Results: In this study, we introduce GCNN-SFM, a computational model for identifying essential genes in organisms, based on graph convolutional neural networks (GCNN). GCNN-SFM integrates a graph convolutional layer, a convolutional layer, and a fully connected layer to model and extract features from gene sequences of essential genes. Initially, the gene sequence is transformed into a feature map using coding techniques. Subsequently, a multi-layer GCN is employed to perform graph convolution operations, effectively capturing both local and global features of the gene sequence. Further feature extraction is performed, followed by integrating convolution and fully-connected layers to generate prediction results for essential genes. The gradient descent algorithm is utilized to iteratively update the cross-entropy loss function, thereby enhancing the accuracy of the prediction results. Meanwhile, model parameters are tuned to determine the optimal parameter combination that yields the best prediction performance during training.
Conclusions: Experimental evaluation demonstrates that GCNN-SFM surpasses various advanced essential gene prediction models and achieves an average accuracy of 94.53%. This study presents a novel and effective approach for identifying essential genes, which has significant implications for biology and genomics research.
Keywords: Bioinformatics; Essential genes; Gene sequences; Graphical convolutional neural networks; Machine learning.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- O’Neill RS, Clark DV. The Drosophila melanogaster septin gene Sep2 has a redundant function with the retrogene Sep5 in imaginal cell proliferation but is essential for oogenesis. Genome. 2013;56(12):753–758. - PubMed
-
- Juhas M, Eberl L, Glass JI. Essence of life: essential genes of minimal genomes. Trends Cell Biol. 2011;21(10):562–568. - PubMed
-
- Juhas M, Reuß DR, Zhu B, Commichau FM. Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering. Microbiology. 2014;160(11):2341–2351. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

