Proteomic Analysis Identifies Distinct Protein Patterns for High Ovulation in FecB Mutant Small Tail Han Sheep Granulosa Cells
- PMID: 38200742
- PMCID: PMC10778137
- DOI: 10.3390/ani14010011
Proteomic Analysis Identifies Distinct Protein Patterns for High Ovulation in FecB Mutant Small Tail Han Sheep Granulosa Cells
Abstract
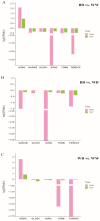
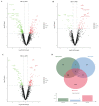
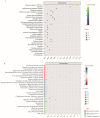
The Booroola fecundity (FecB) mutation in the bone morphogenetic protein receptor type 1B (BMPR1B) gene increases ovulation in sheep. However, its effect on follicular maturation is not fully understood. Therefore, we collected granulosa cells (GCs) at a critical stage of follicle maturation from nine wild-type (WW), nine heterozygous FecB mutant (WB), and nine homozygous FecB mutant (BB) Small Tail Han sheep. The GCs of three ewes were selected at random from each genotype and consolidated into a single group, yielding a total of nine groups (three groups per genotype) for proteomic analysis. The tandem mass tag technique was utilized to ascertain the specific proteins linked to multiple ovulation in the various FecB genotypes. Using a general linear model, we identified 199 proteins significantly affected by the FecB mutation with the LIMMA package (p < 0.05). The differential abundance of proteins was enriched in pathways related to cholesterol metabolism, carbohydrate metabolism, amino acid biosynthesis, and glutathione metabolism. These pathways are involved in important processes for GC-regulated 'conservation' of oocyte maturation. Further, the sparse partial least-squares discriminant analysis and the Fuzzy-C-mean clustering method were combined to estimate weights and cluster differential abundance proteins according to ovulation to screen important ovulation-related proteins. Among them, ZP2 and ZP3 were found to be enriched in the cellular component catalog term "egg coat", as well as some apolipoproteins, such as APOA1, APOA2, and APOA4, enriched in several Gene Ontology terms related to cholesterol metabolism and lipoprotein transport. A higher abundance of these essential proteins for oocyte maturation was observed in BB and WB genotypes compared with WW ewes. These proteins had a high weight in the model for discriminating sheep with different FecB genotypes. These findings provide new insight that the FecB mutant in GCs improves nutrient metabolism, leading to better oocyte maturation by altering the abundance of important proteins (ZP2, ZP3, and APOA1) in favor of increased ovulation or better oocyte quality.
Keywords: FecB mutation; Small Tail Han sheep; fertility; follicular granulosa cells; proteome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mulsant P., Lecerf F., Fabre S., Schibler L., Monget P., Lanneluc I., Pisselet C., Riquet J., Monniaux D., Callebaut I. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Mérino ewes. Proc. Natl. Acad. Sci. USA. 2001;98:5104–5109. doi: 10.1073/pnas.091577598. - DOI - PMC - PubMed
-
- Wilson T., Wu X.Y., Juengel J.L., Ross I.K., Lumsden J.M., Lord E.A., Dodds K.G., Walling G.A., McEwan J.C., O’Connell A.R., et al. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol. Reprod. 2001;64:1225–1235. doi: 10.1095/biolreprod64.4.1225. - DOI - PubMed
-
- Zhou S., Yu H., Zhao X., Cai B., Ding Q., Huang Y., Li Y., Li Y., Niu Y., Lei A., et al. Generation of gene-edited sheep with a defined Booroola fecundity gene (FecBB) mutation in bone morphogenetic protein receptor type 1B (BMPR1B) via clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) 9. Reprod. Fertil. Dev. 2018;30:1616–1621. doi: 10.1071/RD18086. - DOI - PubMed
Grants and funding
- 31861143012/National Natural Science Foundation of China
- 31902150/National Natural Science Foundation of China
- CARS-38/Earmarked Fund for China Agriculture Research System of MOF and MARA
- Y2017JC24/the Central Public-Interest Scientific Institution Basal Research Fund
- 2018ywf-yb-2/Central Public-Interest Scientific Institution Basal Research Fund
LinkOut - more resources
Full Text Sources
Miscellaneous

