Evaluation of Safety and Probiotic Traits from a Comprehensive Genome-Based In Silico Analysis of Ligilactobacillus salivarius P1CEA3, Isolated from Pigs and Producer of Nisin S
- PMID: 38201135
- PMCID: PMC10778751
- DOI: 10.3390/foods13010107
Evaluation of Safety and Probiotic Traits from a Comprehensive Genome-Based In Silico Analysis of Ligilactobacillus salivarius P1CEA3, Isolated from Pigs and Producer of Nisin S
Abstract
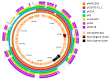
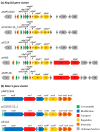
Ligilactobacillus salivarius is an important member of the porcine gastrointestinal tract (GIT). Some L. salivarius strains are considered to have a beneficial effect on the host by exerting different probiotic properties, including the production of antimicrobial peptides which help maintain a healthy gut microbiota. L. salivarius P1CEA3, a porcine isolated strain, was first selected and identified by its antimicrobial activity against a broad range of pathogenic bacteria due to the production of the novel bacteriocin nisin S. The assembled L. salivarius P1CEA3 genome includes a circular chromosome, a megaplasmid (pMP1CEA3) encoding the nisin S gene cluster, and two small plasmids. A comprehensive genome-based in silico analysis of the L. salivarius P1CEA3 genome reveals the presence of genes related to probiotic features such as bacteriocin synthesis, regulation and production, adhesion and aggregation, the production of lactic acid, amino acids metabolism, vitamin biosynthesis, and tolerance to temperature, acid, bile salts and osmotic and oxidative stress. Furthermore, the strain is absent of risk-related genes for acquired antibiotic resistance traits, virulence factors, toxic metabolites and detrimental metabolic or enzymatic activities. Resistance to common antibiotics and gelatinase and hemolytic activities have been discarded by in vitro experiments. This study identifies several probiotic and safety traits of L. salivarius P1CEA3 and suggests its potential as a promising probiotic in swine production.
Keywords: Ligilactobacillus; bacteriocin; megaplasmid; nisin S; probiotic.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Nisin S, a Novel Nisin Variant Produced by Ligilactobacillus salivarius P1CEA3.Int J Mol Sci. 2023 Apr 6;24(7):6813. doi: 10.3390/ijms24076813. Int J Mol Sci. 2023. PMID: 37047785 Free PMC article.
-
Dual bacteriocin and extracellular vesicle-mediated inhibition of Campylobacter jejuni by the potential probiotic candidate Ligilactobacillus salivarius UO.C249.Appl Environ Microbiol. 2024 Aug 21;90(8):e0084524. doi: 10.1128/aem.00845-24. Epub 2024 Jul 30. Appl Environ Microbiol. 2024. PMID: 39078127 Free PMC article.
-
A gut-derived Streptococcus salivarius produces the novel nisin variant designated nisin G and inhibits Fusobacterium nucleatum in a model of the human distal colon microbiome.mBio. 2025 Feb 5;16(2):e0157324. doi: 10.1128/mbio.01573-24. Epub 2024 Dec 18. mBio. 2025. PMID: 39692472 Free PMC article.
-
Understanding Ligilactobacillus salivarius from Probiotic Properties to Omics Technology: A Review.Foods. 2024 Mar 15;13(6):895. doi: 10.3390/foods13060895. Foods. 2024. PMID: 38540885 Free PMC article. Review.
-
Lactobacillus salivarius: bacteriocin and probiotic activity.Food Microbiol. 2013 Dec;36(2):296-304. doi: 10.1016/j.fm.2013.05.010. Epub 2013 Jun 15. Food Microbiol. 2013. PMID: 24010610 Review.
Cited by
-
Screening and Genomic Profiling of Antimicrobial Bacteria Sourced from Poultry Slaughterhouse Effluents: Bacteriocin Production and Safety Evaluation.Genes (Basel). 2024 Dec 2;15(12):1564. doi: 10.3390/genes15121564. Genes (Basel). 2024. PMID: 39766831 Free PMC article.
-
Complete genome sequence, metabolic profiling and functional studies reveal Ligilactobacillus salivarius LS-ARS2 is a promising biofilm-forming probiotic with significant antioxidant, antibacterial, and antibiofilm potential.Front Microbiol. 2025 Mar 20;16:1535388. doi: 10.3389/fmicb.2025.1535388. eCollection 2025. Front Microbiol. 2025. PMID: 40182284 Free PMC article.
-
Therapeutic Potential of Probiotics: A Review of Their Role in Modulating Inflammation.Probiotics Antimicrob Proteins. 2025 Jun 4. doi: 10.1007/s12602-025-10609-z. Online ahead of print. Probiotics Antimicrob Proteins. 2025. PMID: 40465090
-
Distinct phenotypes of salivaricin-producing Ligilactobacillus salivarius isolated from the gastrointestinal tract of broiler chickens and laying hens.Poult Sci. 2025 Jan;104(1):104537. doi: 10.1016/j.psj.2024.104537. Epub 2024 Nov 8. Poult Sci. 2025. PMID: 39571198 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

