Determination of Binding Affinity of Antibodies to HIV-1 Recombinant Envelope Glycoproteins, Pseudoviruses, Infectious Molecular Clones, and Cell-Expressed Trimeric gp160 Using Microscale Thermophoresis
- PMID: 38201237
- PMCID: PMC10778174
- DOI: 10.3390/cells13010033
Determination of Binding Affinity of Antibodies to HIV-1 Recombinant Envelope Glycoproteins, Pseudoviruses, Infectious Molecular Clones, and Cell-Expressed Trimeric gp160 Using Microscale Thermophoresis
Abstract
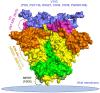
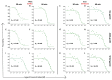
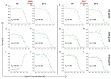
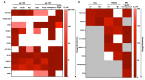
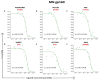
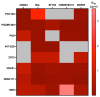
Developing a preventative vaccine for HIV-1 has been a global priority. The elicitation of broadly neutralizing antibodies (bNAbs) against a broad range of HIV-1 envelopes (Envs) from various strains appears to be a critical requirement for an efficacious HIV-1 vaccine. To understand their ability to neutralize HIV-1, it is important to characterize the binding characteristics of bNAbs. Our work is the first to utilize microscale thermophoresis (MST), a rapid, economical, and flexible in-solution temperature gradient method to quantitatively determine the binding affinities of bNAbs and non-neutralizing monoclonal antibodies (mAbs) to HIV-1 recombinant envelope monomer and trimer proteins of different subtypes, pseudoviruses (PVs), infectious molecular clones (IMCs), and cells expressing the trimer. Our results demonstrate that the binding affinities were subtype-dependent. The bNAbs exhibited a higher affinity to IMCs compared to PVs and recombinant proteins. The bNAbs and mAbs bound with high affinity to native-like gp160 trimers expressed on the surface of CEM cells compared to soluble recombinant proteins. Interesting differences were seen with V2-specific mAbs. Although they recognize linear epitopes, one of the antibodies also bound to the Envs on PVs, IMCs, and a recombinant trimer protein, suggesting that the epitope was not occluded. The identification of epitopes on the envelope surface that can bind to high affinity mAbs could be useful for designing HIV-1 vaccines and for down-selecting vaccine candidates that can induce high affinity antibodies to the HIV-1 envelope in their native conformation.
Keywords: HIV-1 epitopes; MST; bNAb; binding affinity; cell-expressed trimeric protein; infectious molecular clone; mAb; pseudovirus; recombinant HIV-1 proteins.
Conflict of interest statement
S.B., N.G., J.K., H.V.T., M.C., M.G.J. employed by The Henry M. Jackson Foundation for the Advancement of Military Medicine. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures

References
-
- UNAIDS. 2023. [(accessed on 4 October 2023)]. Available online: https://www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_....
-
- Fouts T., Godfrey K., Bobb K., Montefiori D., Hanson C.V., Kalyanaraman V.S., DeVico A., Pal R. Crosslinked HIV-1 envelope–CD4 receptor complexes elicit broadly cross-reactive neutralizing antibodies in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2002;99:11842–11847. doi: 10.1073/pnas.182412199. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

