Metabolic Changes during In Vivo Maturation of PSC-Derived Skeletal Myogenic Progenitors
- PMID: 38201280
- PMCID: PMC10778145
- DOI: 10.3390/cells13010076
Metabolic Changes during In Vivo Maturation of PSC-Derived Skeletal Myogenic Progenitors
Abstract
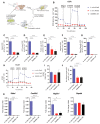
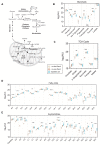
In vitro-generated pluripotent stem cell (PSC)-derived Pax3-induced (iPax3) myogenic progenitors display an embryonic transcriptional signature, but upon engraftment, the profile of re-isolated iPax3 donor-derived satellite cells changes toward similarity with postnatal satellite cells, suggesting that engrafted PSC-derived myogenic cells remodel their transcriptional signature upon interaction within the adult muscle environment. Here, we show that engrafted myogenic progenitors also remodel their metabolic state. Assessment of oxygen consumption revealed that exposure to the adult muscle environment promotes overt changes in mitochondrial bioenergetics, as shown by the substantial suppression of energy requirements in re-isolated iPax3 donor-derived satellite cells compared to their in vitro-generated progenitors. Mass spectrometry-based metabolomic profiling further confirmed the relationship of engrafted iPax3 donor-derived cells to adult satellite cells. The fact that in vitro-generated myogenic progenitors remodel their bioenergetic signature upon in vivo exposure to the adult muscle environment may have important implications for therapeutic applications.
Keywords: in vitro; in vivo; metabolism; myogenic progenitors; pluripotent stem cells; satellite cells.
Conflict of interest statement
R.C.R.P. is cofounder and holds equity in Myogenica. T.N. is a cofounder and holds equity in Omix Technologies. All other authors have no competing financial interests.
Figures


References
-
- Ikemoto M., Fukada S., Uezumi A., Masuda S., Miyoshi H., Yamamoto H., Wada M.R., Masubuchi N., Miyagoe-Suzuki Y., Takeda S. Autologous transplantation of SM/C-2.6(+) satellite cells transduced with micro-dystrophin CS1 cDNA by lentiviral vector into mdx mice. Mol. Ther. 2007;15:2178–2185. doi: 10.1038/sj.mt.6300295. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

