High Mitochondrial Protein Expression as a Potential Predictor of Relapse Risk in Acute Myeloid Leukemia Patients with the Monocytic FAB Subtypes M4 and M5
- PMID: 38201437
- PMCID: PMC10778527
- DOI: 10.3390/cancers16010008
High Mitochondrial Protein Expression as a Potential Predictor of Relapse Risk in Acute Myeloid Leukemia Patients with the Monocytic FAB Subtypes M4 and M5
Abstract
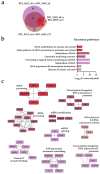
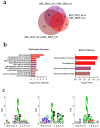
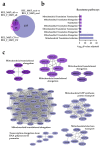
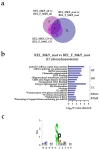
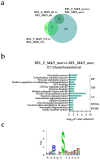
AML is a highly aggressive and heterogeneous form of hematological cancer. Proteomics-based stratification of patients into more refined subgroups may contribute to a more precise characterization of the patient-derived AML cells. Here, we reanalyzed liquid chromatography-tandem mass spectrometry (LC-MS/MS) generated proteomic and phosphoproteomic data from 26 FAB-M4/M5 patients. The patients achieved complete hematological remission after induction therapy. Twelve of them later developed chemoresistant relapse (RELAPSE), and 14 patients were relapse-free (REL_FREE) long-term survivors. We considered not only the RELAPSE and REL_FREE characteristics but also integrated the French-American-British (FAB) classification, along with considering the presence of nucleophosmin 1 (NPM1) mutation and cytogenetically normal AML. We found a significant number of differentially enriched proteins (911) and phosphoproteins (257) between the various FAB subtypes in RELAPSE patients. Patients with the myeloblastic M1/M2 subtype showed higher levels of RNA processing-related routes and lower levels of signaling related to terms like translation and degranulation when compared with the M4/M5 subtype. Moreover, we found that a high abundance of proteins associated with mitochondrial translation and oxidative phosphorylation, particularly observed in the RELAPSE M4/M5 NPM1 mutated subgroup, distinguishes relapsing from non-relapsing AML patient cells with the FAB subtype M4/M5. Thus, the discovery of subtype-specific biomarkers through proteomic profiling may complement the existing classification system for AML and potentially aid in selecting personalized treatment strategies for individual patients.
Keywords: FAB subtypes; acute myeloid leukemia; mass spectrometry; mitochondria; phosphoproteomic; proteomic; relapse.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Dohner H., Wei A.H., Appelbaum F.R., Craddock C., DiNardo C.D., Dombret H., Ebert B.L., Fenaux P., Godley L.A., Hasserjian R.P., et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–1377. doi: 10.1182/blood.2022016867. - DOI - PubMed
-
- Khoury J.D., Solary E., Abla O., Akkari Y., Alaggio R., Apperley J.F., Bejar R., Berti E., Busque L., Chan J.K.C., et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 2022;36:1703–1719. doi: 10.1038/s41375-022-01613-1. - DOI - PMC - PubMed
-
- Dohner H., Estey E.H., Amadori S., Appelbaum F.R., Buchner T., Burnett A.K., Dombret H., Fenaux P., Grimwade D., Larson R.A., et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. - DOI - PubMed
-
- Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. - DOI - PubMed
LinkOut - more resources
Full Text Sources

