Implementing an On-Slide Molecular Classification of Gastric Cancer: A Tissue Microarray Study
- PMID: 38201483
- PMCID: PMC10778243
- DOI: 10.3390/cancers16010055
Implementing an On-Slide Molecular Classification of Gastric Cancer: A Tissue Microarray Study
Abstract
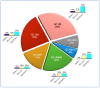
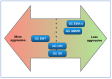
Background and Objectives: Gastric cancer (GC) is one of the most commonly diagnosed cancers and the fourth cause of cancer death worldwide. Personalised treatment improves GC outcomes. A molecular classification is needed to choose the appropriate therapy. A classification that uses on-slide biomarkers and formalin-fixed and paraffin-embedded (FFPE) tissue is preferable to comprehensive genomic analysis. In 2016, Setia and colleagues proposed an on-slide classification; however, this is not in widespread use. We propose a modification of this classification that has six subgroups: GC associated with Epstein-Barr virus (GC EBV+), GC with mismatch-repair deficiency (GC dMMR), GC with epithelial-mesenchymal transformation (GC EMT), GC with chromosomal instability (GC CIN), CG that is genomically stable (GC GS) and GC not otherwise specified (GC NOS). This classification also has a provision for biomarkers for current or emerging targeted therapies (Her2, PD-L1 and Claudin18.2). Here, we assess the implementation and feasibility of this inclusive working classification. Materials and Methods: We constructed a tissue microarray library from a cohort of 79 resection cases from FFPE tissue archives. We used a restricted panel of on-slide markers (EBER, MMR, E-cadherin, beta-catenin and p53), defined their interpretation algorithms and assigned each case to a specific molecular subtype. Results: GC EBV(+) cases were 6%, GC dMMR cases were 20%, GC EMT cases were 14%, GC CIN cases were 23%, GC GS cases were 29%, and GC NOS cases were 8%. Conclusions: This working classification uses markers that are widely available in histopathology and are easy to interpret. A diagnostic subgroup is obtained for 92% of the cases. The proportion of cases in each subgroup is in keeping with other published series. Widescale implementation appears feasible. A study using endoscopic biopsies is warranted.
Keywords: Claudin18.2; E-cadherin; EBER; Her2; MMR; PD-L1; beta-catenin; gastric cancer; molecular classification; p53.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Implementing an integrated molecular classification for gastric cancer from endoscopic biopsies using on-slide tests.Rom J Morphol Embryol. 2024 Apr-Jun;65(2):257-265. doi: 10.47162/RJME.65.2.12. Rom J Morphol Embryol. 2024. PMID: 39020540 Free PMC article.
-
Prevalence of Epstein-Barr Virus Infection and Mismatch Repair Protein Deficiency and the Correlation of Immune Markers in Tibetan Patients with Gastric Cancer.Biomed Res Int. 2022 Jun 13;2022:2684065. doi: 10.1155/2022/2684065. eCollection 2022. Biomed Res Int. 2022. PMID: 35734348 Free PMC article.
-
Development and Validation of an Easy-to-Implement, Practical Algorithm for the Identification of Molecular Subtypes of Gastric Cancer: Prognostic and Therapeutic Implications.Oncologist. 2019 Dec;24(12):e1321-e1330. doi: 10.1634/theoncologist.2019-0058. Epub 2019 Aug 1. Oncologist. 2019. PMID: 31371521 Free PMC article.
-
A consolidated working classification of gastric cancer for histopathologists (Review).Biomed Rep. 2023 Jul 19;19(3):58. doi: 10.3892/br.2023.1640. eCollection 2023 Sep. Biomed Rep. 2023. PMID: 37614984 Free PMC article. Review.
-
Chromosomal Instability in Gastric Cancer: Role in Tumor Development, Progression, and Therapy.Int J Mol Sci. 2023 Nov 30;24(23):16961. doi: 10.3390/ijms242316961. Int J Mol Sci. 2023. PMID: 38069284 Free PMC article. Review.
Cited by
-
Implementing an integrated molecular classification for gastric cancer from endoscopic biopsies using on-slide tests.Rom J Morphol Embryol. 2024 Apr-Jun;65(2):257-265. doi: 10.47162/RJME.65.2.12. Rom J Morphol Embryol. 2024. PMID: 39020540 Free PMC article.
-
HtrA-Dependent E-Cadherin Shedding Impairs the Epithelial Barrier Function in Primary Gastric Epithelial Cells and Gastric Organoids.Int J Mol Sci. 2024 Jun 27;25(13):7083. doi: 10.3390/ijms25137083. Int J Mol Sci. 2024. PMID: 39000189 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

