A Novel Tissue-Free Method to Estimate Tumor-Derived Cell-Free DNA Quantity Using Tumor Methylation Patterns
- PMID: 38201510
- PMCID: PMC10777919
- DOI: 10.3390/cancers16010082
A Novel Tissue-Free Method to Estimate Tumor-Derived Cell-Free DNA Quantity Using Tumor Methylation Patterns
Abstract
Estimating the abundance of cell-free DNA (cfDNA) fragments shed from a tumor (i.e., circulating tumor DNA (ctDNA)) can approximate tumor burden, which has numerous clinical applications. We derived a novel, broadly applicable statistical method to quantify cancer-indicative methylation patterns within cfDNA to estimate ctDNA abundance, even at low levels. Our algorithm identified differentially methylated regions (DMRs) between a reference database of cancer tissue biopsy samples and cfDNA from individuals without cancer. Then, without utilizing matched tissue biopsy, counts of fragments matching the cancer-indicative hyper/hypo-methylated patterns within DMRs were used to determine a tumor methylated fraction (TMeF; a methylation-based quantification of the circulating tumor allele fraction and estimate of ctDNA abundance) for plasma samples. TMeF and small variant allele fraction (SVAF) estimates of the same cancer plasma samples were correlated (Spearman's correlation coefficient: 0.73), and synthetic dilutions to expected TMeF of 10-3 and 10-4 had estimated TMeF within two-fold for 95% and 77% of samples, respectively. TMeF increased with cancer stage and tumor size and inversely correlated with survival probability. Therefore, tumor-derived fragments in the cfDNA of patients with cancer can be leveraged to estimate ctDNA abundance without the need for a tumor biopsy, which may provide non-invasive clinical approximations of tumor burden.
Keywords: DNA methylation; biomarkers; cell-free DNA; circulating tumor DNA; computational methods; liquid biopsy; machine learning algorithms; tumor fraction; variant allele fraction.
Conflict of interest statement
Y.Z., M.R.-S. and A.H.S. are employees of GRAIL, LLC. C.C., S.S. and G.C. are employees of GRAIL, LLC with equity in Illumina, Inc. C.A.M., A.S., C.C.K., E.S. and P.-Y.C. are employees of GRAIL, LLC with equity in GRAIL, LLC and Illumina, Inc. P.F. and S.B. were previously employed by GRAIL, LLC with equity in GRAIL, LLC and Illumina, Inc.
Figures
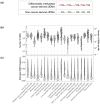
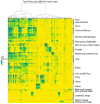
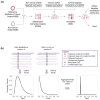

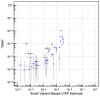
References
-
- Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1) Eur. J. Cancer Oxf. Engl. 1990. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

