AI-Powered Segmentation of Invasive Carcinoma Regions in Breast Cancer Immunohistochemical Whole-Slide Images
- PMID: 38201594
- PMCID: PMC10778369
- DOI: 10.3390/cancers16010167
AI-Powered Segmentation of Invasive Carcinoma Regions in Breast Cancer Immunohistochemical Whole-Slide Images
Abstract
Aims: The automation of quantitative evaluation for breast immunohistochemistry (IHC) plays a crucial role in reducing the workload of pathologists and enhancing the objectivity of diagnoses. However, current methods face challenges in achieving fully automated immunohistochemistry quantification due to the complexity of segmenting the tumor area into distinct ductal carcinoma in situ (DCIS) and invasive carcinoma (IC) regions. Moreover, the quantitative analysis of immunohistochemistry requires a specific focus on invasive carcinoma regions.
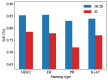
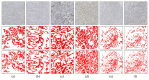
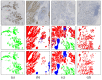
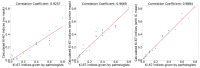
Methods and results: In this study, we propose an innovative approach to automatically identify invasive carcinoma regions in breast cancer immunohistochemistry whole-slide images (WSIs). Our method leverages a neural network that combines multi-scale morphological features with boundary features, enabling precise segmentation of invasive carcinoma regions without the need for additional H&E and P63 staining slides. In addition, we introduced an advanced semi-supervised learning algorithm, allowing efficient training of the model using unlabeled data. To evaluate the effectiveness of our approach, we constructed a dataset consisting of 618 IHC-stained WSIs from 170 cases, including four types of staining (ER, PR, HER2, and Ki-67). Notably, the model demonstrated an impressive intersection over union (IoU) score exceeding 80% on the test set. Furthermore, to ascertain the practical utility of our model in IHC quantitative evaluation, we constructed a fully automated Ki-67 scoring system based on the model's predictions. Comparative experiments convincingly demonstrated that our system exhibited high consistency with the scores given by experienced pathologists.
Conclusions: Our developed model excels in accurately distinguishing between DCIS and invasive carcinoma regions in breast cancer immunohistochemistry WSIs. This method paves the way for a clinically available, fully automated immunohistochemistry quantitative scoring system.
Keywords: IHC quantification; Ki-67; artificial intelligence; breast cancer; invasive carcinoma.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Precision HER2: a comprehensive AI system for accurate and consistent evaluation of HER2 expression in invasive breast Cancer.BMC Cancer. 2024 Sep 30;24(1):1204. doi: 10.1186/s12885-024-12980-6. BMC Cancer. 2024. PMID: 39350085 Free PMC article.
-
AutoIHC-Analyzer: computer-assisted microscopy for automated membrane extraction/scoring in HER2 molecular markers.J Microsc. 2021 Jan;281(1):87-96. doi: 10.1111/jmi.12955. Epub 2020 Aug 27. J Microsc. 2021. PMID: 32803890
-
Fully Automated Artificial Intelligence Solution for Human Epidermal Growth Factor Receptor 2 Immunohistochemistry Scoring in Breast Cancer: A Multireader Study.JCO Precis Oncol. 2024 Oct;8:e2400353. doi: 10.1200/PO.24.00353. Epub 2024 Oct 11. JCO Precis Oncol. 2024. PMID: 39393036 Free PMC article.
-
Automated quantitative analysis of Ki-67 staining and HE images recognition and registration based on whole tissue sections in breast carcinoma.Diagn Pathol. 2020 May 29;15(1):65. doi: 10.1186/s13000-020-00957-5. Diagn Pathol. 2020. PMID: 32471471 Free PMC article.
-
HoLy-Net: Segmentation of histological images of diffuse large B-cell lymphoma.Comput Biol Med. 2024 Mar;170:107978. doi: 10.1016/j.compbiomed.2024.107978. Epub 2024 Jan 11. Comput Biol Med. 2024. PMID: 38237235
Cited by
-
Carvedilol sensitizes paclitaxel-resistant gastric cancer AGS cells to paclitaxel: influences on apoptotic regulators, Notch, PI3K/AKT, ERK1/2 signaling pathways, and miR-34a expression.Med Oncol. 2025 Jul 30;42(9):392. doi: 10.1007/s12032-025-02966-0. Med Oncol. 2025. PMID: 40736757
-
The role of HOXA1 in cancer and targeted therapy.Med Oncol. 2025 Aug 4;42(9):405. doi: 10.1007/s12032-025-02980-2. Med Oncol. 2025. PMID: 40760270 Review.
-
Mesenchymal stem/stromal cells: dedicator to maintain tumor homeostasis.Hum Cell. 2024 Nov 28;38(1):21. doi: 10.1007/s13577-024-01154-y. Hum Cell. 2024. PMID: 39607530 Review.
-
KDM1A-mediated ZFP64 demethylation activates CENPL to promote epithelial ovarian cancer progression.Cytotechnology. 2025 Feb;77(1):10. doi: 10.1007/s10616-024-00671-w. Epub 2024 Dec 1. Cytotechnology. 2025. PMID: 39628712
-
Extracellular vesicles derived from bone marrow mesenchymal stem cells ameliorate chronic liver damage via microRNA-136-5p.Mol Cell Biochem. 2025 Feb;480(2):951-969. doi: 10.1007/s11010-024-04993-3. Epub 2024 Apr 23. Mol Cell Biochem. 2025. PMID: 38652214
References
-
- Chhikara B.S., Parang K. Global Cancer Statistics 2022: The trends projection analysis. Chem. Biol. Lett. 2023;10:451.
-
- WHO . WHO Classification of Tumors–Breast Tumors. 5th ed. International Agency for Research on Cancer; Lyon, France: 2019.
-
- Dabbs D.J. Diagnostic Immunohistochemistry E-Book: Theranostic and Genomic Applications. Elsevier; Amsterdam, The Netherlands: 2021.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

