Identifying and Characterizing Candidate Genes Contributing to a Grain Yield QTL in Wheat
- PMID: 38202333
- PMCID: PMC10780351
- DOI: 10.3390/plants13010026
Identifying and Characterizing Candidate Genes Contributing to a Grain Yield QTL in Wheat
Abstract
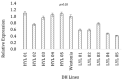
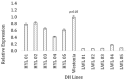
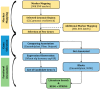
The current study focuses on identifying the candidate genes of a grain yield QTL from a double haploid population, Westonia × Kauz. The QTL region spans 20 Mbp on the IWGSC whole-genome sequence flank with 90K SNP markers. The IWGSC gene annotation revealed 16 high-confidence genes and 41 low-confidence genes. Bioinformatic approaches, including functional gene annotation, ontology investigation, pathway exploration, and gene network study using publicly available gene expression data, enabled the short-listing of four genes for further confirmation. Complete sequencing of those four genes demonstrated that only two genes are polymorphic between the parental cultivars, which are the ferredoxin-like protein gene and the tetratricopeptide-repeat (TPR) protein gene. The two genes were selected for downstream investigation. Two SNP variations were observed in the exon for both genes, with one SNP resulting in changes in amino acid sequence. qPCR-based gene expression showed that both genes were highly expressed in the high-yielding double haploid lines along with the parental cultivar Westonia. In contrast, their expression was significantly lower in the low-yielding lines in the other parent. It can be concluded that these two genes are the contributing genes to the grain yield QTL.
Keywords: bioinformatics; gene identification; quantitative trait locus (QTL).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Development of a High-Density SNP-Based Linkage Map and Detection of QTL for β-Glucans, Protein Content, Grain Yield per Spike and Heading Time in Durum Wheat.Int J Mol Sci. 2017 Jun 21;18(6):1329. doi: 10.3390/ijms18061329. Int J Mol Sci. 2017. PMID: 28635630 Free PMC article.
-
QTL mapping for grain yield and three yield components in a population derived from two high-yielding spring wheat cultivars.Theor Appl Genet. 2021 Jul;134(7):2079-2095. doi: 10.1007/s00122-021-03806-1. Epub 2021 Mar 9. Theor Appl Genet. 2021. PMID: 33687497 Free PMC article.
-
Identification of candidate genes, regions and markers for pre-harvest sprouting resistance in wheat (Triticum aestivum L.).BMC Plant Biol. 2014 Nov 29;14:340. doi: 10.1186/s12870-014-0340-1. BMC Plant Biol. 2014. PMID: 25432597 Free PMC article.
-
Exotic QTL improve grain quality in the tri-parental wheat population SW84.PLoS One. 2017 Jul 7;12(7):e0179851. doi: 10.1371/journal.pone.0179851. eCollection 2017. PLoS One. 2017. PMID: 28686676 Free PMC article.
-
Using genetic mapping and genomics approaches in understanding and improving drought tolerance in pearl millet.J Exp Bot. 2011 Jan;62(2):397-408. doi: 10.1093/jxb/erq265. Epub 2010 Sep 5. J Exp Bot. 2011. PMID: 20819788 Review.
References
-
- Abdurakhmonov I.Y. Bioinformatics: Basics, Development, and Future. IntechOpen; London, UK: 2016. Bioinformatics—Updated features and applications. - DOI
-
- Adamski N.M., Borrill P., Brinton J., Harrington S.A., Marchal C., Bentley A.R., Bovill W.D., Cattivelli L., Cockram J., Moreira B.C., et al. eLife. Volume 9. eLife Sciences Publications Ltd.; Cambridge, UK: 2020. A roadmap for gene functional characterisation in crops with large genomes: Lessons from polyploid wheat. - DOI - PMC - PubMed
-
- An D., Su J., Liu Q., Zhu Y., Tong Y., Li J., Jing R., Li B., Li Z. Mapping QTLs for nitrogen uptake in relation to the early growth of wheat (Triticum aestivum L.) Plant Soil. 2006;284:73–84. doi: 10.1007/s11104-006-0030-3. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

