NLR- and mlo-Based Resistance Mechanisms against Powdery Mildew in Cannabis sativa
- PMID: 38202413
- PMCID: PMC10780410
- DOI: 10.3390/plants13010105
NLR- and mlo-Based Resistance Mechanisms against Powdery Mildew in Cannabis sativa
Abstract
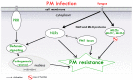
Powdery mildew (PM) is one of the most common Cannabis sativa diseases. In spite of this, very few documented studies have characterized the resistance genes involved in PM defense mechanisms, or sources of natural genetic resistance in cannabis. The focus of the present work is on the two primary mechanisms for qualitative resistance against PM. The first is based on resistance (R) genes characterized by conserved nucleotide-binding site and/or leucine-rich repeat domains (NLRs). The second one involves susceptibility (S) genes, and particularly mildew resistance locus o (MLO) genes, whose loss-of-function mutations seem to be a reliable way to protect plants from PM infection. Cannabis defenses against PM are thus discussed, mainly detailing the strategies based on these two mechanisms. Emerging studies about this research topic are also reported and, based on the most significant results, a potential PM resistance model in cannabis plant-pathogen interactions is proposed. Finally, innovative approaches, based on the pyramiding of multiple R genes, as well as on genetic engineering and genome editing methods knocking out S genes, are discussed, to obtain durable PM-resistant cannabis cultivars with a broad-spectrum resistance range.
Keywords: Cannabis sativa; broad-spectrum resistance; disease resistance genes; mildew resistance locus o; nucleotide-binding and leucine-rich repeat receptors; powdery mildew.
Conflict of interest statement
The author declares no conflicts of interest.
Figures
Similar articles
-
Mapping and characterization of a novel powdery mildew resistance locus (PM2) in Cannabis sativa L.Front Plant Sci. 2025 Mar 13;16:1543229. doi: 10.3389/fpls.2025.1543229. eCollection 2025. Front Plant Sci. 2025. PMID: 40182551 Free PMC article.
-
Genome-Wide Characterization of the MLO Gene Family in Cannabis sativa Reveals Two Genes as Strong Candidates for Powdery Mildew Susceptibility.Front Plant Sci. 2021 Sep 13;12:729261. doi: 10.3389/fpls.2021.729261. eCollection 2021. Front Plant Sci. 2021. PMID: 34589104 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
Cited by
-
Integrated Management of Pathogens and Microbes in Cannabis sativa L. (Cannabis) under Greenhouse Conditions.Plants (Basel). 2024 Mar 10;13(6):786. doi: 10.3390/plants13060786. Plants (Basel). 2024. PMID: 38592798 Free PMC article. Review.
-
Mapping and characterization of a novel powdery mildew resistance locus (PM2) in Cannabis sativa L.Front Plant Sci. 2025 Mar 13;16:1543229. doi: 10.3389/fpls.2025.1543229. eCollection 2025. Front Plant Sci. 2025. PMID: 40182551 Free PMC article.
-
Transcriptional time-course analysis during ash dieback infection revealed different responses in tolerant and susceptible Fraxinus excelsior genotypes.BMC Plant Biol. 2025 Jan 25;25(1):107. doi: 10.1186/s12870-025-06074-z. BMC Plant Biol. 2025. PMID: 39856539 Free PMC article.
-
Major Genes for Powdery Mildew Resistance in Research and Breeding of Barley: A Few Brief Narratives and Recommendations.Plants (Basel). 2025 Jul 8;14(14):2091. doi: 10.3390/plants14142091. Plants (Basel). 2025. PMID: 40733328 Free PMC article. Review.
-
Reduction of flavonoid content in honeysuckle via Erysiphe lonicerae-mediated inhibition of three essential genes in flavonoid biosynthesis pathways.Front Plant Sci. 2024 Apr 16;15:1381368. doi: 10.3389/fpls.2024.1381368. eCollection 2024. Front Plant Sci. 2024. PMID: 38689843 Free PMC article.
References
-
- Chandran D., Wildermuth M.C. Modulation of Host Endocycle During Plant–Biotroph Interactions. Enzymes. 2016;40:65–103. - PubMed
-
- Hacquard S. The Genomics of Powdery Mildew Fungi: Past Achievements, Present Status and Future Prospects. Adv. Bot. Res. 2014;70:109–142.
-
- Keinath A.P., DuBose V.B. Controlling powdery mildew on cucurbit rootstock seedlings in the greenhouse with fungicides and biofungicides. Crop Prot. 2012;42:338–344. doi: 10.1016/j.cropro.2012.06.009. - DOI
Publication types
LinkOut - more resources
Full Text Sources

