Comparison of Toxicity and Cellular Uptake of CdSe/ZnS and Carbon Quantum Dots for Molecular Tracking Using Saccharomyces cerevisiae as a Fungal Model
- PMID: 38202465
- PMCID: PMC10781119
- DOI: 10.3390/nano14010010
Comparison of Toxicity and Cellular Uptake of CdSe/ZnS and Carbon Quantum Dots for Molecular Tracking Using Saccharomyces cerevisiae as a Fungal Model
Abstract
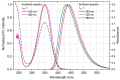
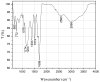
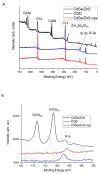
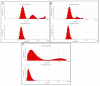
Plant resource sharing mediated by mycorrhizal fungi has been a subject of recent debate, largely owing to the limitations of previously used isotopic tracking methods. Although CdSe/ZnS quantum dots (QDs) have been successfully used for in situ tracking of essential nutrients in plant-fungal systems, the Cd-containing QDs, due to the intrinsic toxic nature of Cd, are not a viable system for larger-scale in situ studies. We synthesized amino acid-based carbon quantum dots (CQDs; average hydrodynamic size 6 ± 3 nm, zeta potential -19 ± 12 mV) and compared their toxicity and uptake with commercial CdSe/ZnS QDs that we conjugated with the amino acid cysteine (Cys) (average hydrodynamic size 308 ± 150 nm, zeta potential -65 ± 4 mV) using yeast Saccharomyces cerevisiae as a proxy for mycorrhizal fungi. We showed that the CQDs readily entered yeast cells and were non-toxic up to 100 mg/L. While the Cys-conjugated CdSe/ZnS QDs were also not toxic to yeast cells up to 100 mg/L, they were not taken up into the cells but remained on the cell surfaces. These findings suggest that CQDs may be a suitable tool for molecular tracking in fungi (incl. mychorrhizal fungi) due to their ability to enter fungal cells.
Keywords: Saccharomyces cerevisiae; carbon dots; molecular tracking; mycorrhizal fungi; nanoparticle toxicity; nanoparticle uptake; nutrients; quantum dots; viability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The interactions between CdSe quantum dots and yeast Saccharomyces cerevisiae: adhesion of quantum dots to the cell surface and the protection effect of ZnS shell.Chemosphere. 2014 Oct;112:92-9. doi: 10.1016/j.chemosphere.2014.03.071. Epub 2014 Apr 21. Chemosphere. 2014. PMID: 25048893
-
Transcriptome Profile Alteration with Cadmium Selenide/Zinc Sulfide Quantum Dots in Saccharomyces cerevisiae.Biomolecules. 2019 Oct 25;9(11):653. doi: 10.3390/biom9110653. Biomolecules. 2019. PMID: 31731522 Free PMC article.
-
Mitigating the toxic effects of CdSe quantum dots towards freshwater alga Scenedesmus obliquus: Role of eco-corona.Environ Pollut. 2021 Feb 1;270:116049. doi: 10.1016/j.envpol.2020.116049. Epub 2020 Nov 10. Environ Pollut. 2021. PMID: 33213955
-
Cellular uptake, elimination and toxicity of CdSe/ZnS quantum dots in HepG2 cells.Biomaterials. 2013 Dec;34(37):9545-58. doi: 10.1016/j.biomaterials.2013.08.038. Epub 2013 Sep 5. Biomaterials. 2013. PMID: 24011712
-
The Impact of Cadmium Selenide Zinc Sulfide Quantum Dots on the Proteomic Profile of Saccharomyces cerevisiae.Int J Mol Sci. 2023 Nov 15;24(22):16332. doi: 10.3390/ijms242216332. Int J Mol Sci. 2023. PMID: 38003523 Free PMC article.
Cited by
-
Cytotoxicity of Quantum Dots in Receptor-Mediated Endocytic and Pinocytic Pathways in Yeast.Int J Mol Sci. 2024 Apr 26;25(9):4714. doi: 10.3390/ijms25094714. Int J Mol Sci. 2024. PMID: 38731933 Free PMC article.
-
Carbon Dots as an Emergent Class of Sustainable Antifungal Agents.ACS Nano. 2025 Jul 15;19(27):24377-24403. doi: 10.1021/acsnano.5c03934. Epub 2025 Jul 2. ACS Nano. 2025. PMID: 40601415 Free PMC article. Review.
References
-
- Hevesy G. Application of Isotopes in Biology. J. Chem. Soc. 1939:1213–1223. doi: 10.1039/jr9390001213. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

