Diversity of Self-Assembled RNA Complexes: From Nanoarchitecture to Nanomachines
- PMID: 38202593
- PMCID: PMC10779776
- DOI: 10.3390/molecules29010010
Diversity of Self-Assembled RNA Complexes: From Nanoarchitecture to Nanomachines
Abstract
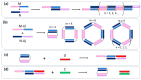
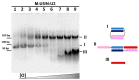
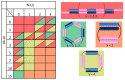
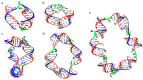
New tool development for various nucleic acid applications is an essential task in RNA nanotechnology. Here, we determined the ability of self-limited complex formation by a pair of oligoribonucleotides carrying two pairwise complementary blocks connected by a linker of different lengths in each chain. The complexes were analyzed using UV melting, gel shift assay analysis, and molecular dynamics (MD) simulations. We have demonstrated the spontaneous formation of various self-limited and concatemer complexes. The linear concatemer complex is formed by a pair of oligomers without a linker in at least one of them. Longer linkers resulted in the formation of circular complexes. The self-limited complexes formation was confirmed using the toehold strand displacement. The MD simulations indicate the reliability of the complexes' structure and demonstrate their dynamics, which increase with the rise of complex size. The linearization of 2D circular complexes into 1D structures and a reverse cyclization process were demonstrated using a toehold-mediated approach. The approach proposed here for the construction and directed modification of the molecularity and shape of complexes will be a valuable tool in RNA nanotechnology, especially for the rational design of therapeutic nucleic acids with high target specificity and the programmable response of the immune system of organisms.
Keywords: RNA; circular RNA; concatemer; molecular dynamics of nucleic acids; rational design; self-assembly; self-limited complex; supramolecular complexes of nucleic acids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Hybrid RNA/DNA Concatemers and Self-Limited Complexes: Structure and Prospects for Therapeutic Applications.Molecules. 2024 Dec 13;29(24):5896. doi: 10.3390/molecules29245896. Molecules. 2024. PMID: 39769985 Free PMC article.
-
Pairing nanoarchitectonics of oligodeoxyribonucleotides with complex diversity: concatemers and self-limited complexes.RSC Adv. 2022 Feb 23;12(11):6416-6431. doi: 10.1039/d2ra00155a. eCollection 2022 Feb 22. RSC Adv. 2022. PMID: 35424594 Free PMC article.
-
Toehold clipping: A mechanism for remote control of DNA strand displacement.Nucleic Acids Res. 2023 May 8;51(8):4055-4063. doi: 10.1093/nar/gkac1152. Nucleic Acids Res. 2023. PMID: 36477864 Free PMC article.
-
Lipophilic nucleic acids--a flexible construction kit for organization and functionalization of surfaces.Adv Colloid Interface Sci. 2014 Jun;208:235-51. doi: 10.1016/j.cis.2014.02.019. Epub 2014 Mar 5. Adv Colloid Interface Sci. 2014. PMID: 24650567 Review.
-
Principles of nucleic acid toehold mediated strand displacement (TMSD) reaction model and its applications in cell environment.Biomater Sci. 2023 Jul 25;11(15):5060-5077. doi: 10.1039/d3bm00476g. Biomater Sci. 2023. PMID: 37260180 Review.
Cited by
-
Hybrid RNA/DNA Concatemers and Self-Limited Complexes: Structure and Prospects for Therapeutic Applications.Molecules. 2024 Dec 13;29(24):5896. doi: 10.3390/molecules29245896. Molecules. 2024. PMID: 39769985 Free PMC article.
-
Inhibition of SARS-CoV-2 Replication by Self-Assembled siRNA Nanoparticles Targeting Multiple Highly Conserved Viral Sequences.Viruses. 2024 Jul 3;16(7):1072. doi: 10.3390/v16071072. Viruses. 2024. PMID: 39066234 Free PMC article.
References
-
- Binzel D.W., Li X., Burns N., Khan E., Lee W.J., Chen L.C., Ellipilli S., Miles W., Ho Y.S., Guo P. Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine-Specific Cancer Targeting with Undetectable Toxicity. Chem. Rev. 2021;121:7398–7467. doi: 10.1021/acs.chemrev.1c00009. - DOI - PMC - PubMed
-
- Green A.A. RNA Nanostructures. Volume 1632. Humana; New York, NY, USA: 2017. pp. 285–302. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

