Design and Synthesis of Novel Indole Ethylamine Derivatives as a Lipid Metabolism Regulator Targeting PPARα/CPT1 in AML12 Cells
- PMID: 38202597
- PMCID: PMC10779794
- DOI: 10.3390/molecules29010012
Design and Synthesis of Novel Indole Ethylamine Derivatives as a Lipid Metabolism Regulator Targeting PPARα/CPT1 in AML12 Cells
Abstract
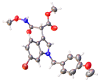
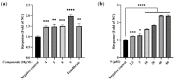
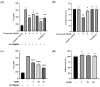
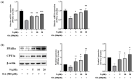
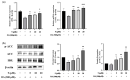
Peroxisome proliferator-activated receptor alpha (PPARα) and carnitine palmitoyltransferase 1 (CPT1) are important targets of lipid metabolism regulation for nonalcoholic fatty liver disease (NAFLD) therapy. In the present study, a set of novel indole ethylamine derivatives (4, 5, 8, 9) were designed and synthesized. The target product (compound 9) can effectively activate PPARα and CPT1a. Consistently, in vitro assays demonstrated its impact on the lipid accumulation of oleic acid (OA)-induced AML12 cells. Compared with AML12 cells treated only with OA, supplementation with 5, 10, and 20 μM of compound 9 reduced the levels of intracellular triglyceride (by 28.07%, 37.55%, and 51.33%) with greater inhibitory activity relative to the commercial PPARα agonist fenofibrate. Moreover, the compound 9 supplementations upregulated the expression of hormone-sensitive triglyceride lipase (HSL) and adipose triglyceride lipase (ATGL) and upregulated the phosphorylation of acetyl-CoA carboxylase (ACC) related to fatty acid oxidation and lipogenesis. This dual-target compound with lipid metabolism regulatory efficacy may represent a promising type of drug lead for NAFLD therapy.
Keywords: CPT1; NAFLD; PPARα; indole ethylamine derivatives.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Chiriac S., Stanciu C., Girleanu I., Cojocariu C., Sfarti C., Singeap A.M., Cuciureanu T., Huiban L., Muzica C.M., Zenovia S., et al. Nonalcoholic Fatty Liver Disease and Cardiovascular Diseases: The Heart of the Matter. Can. J. Gastroenterol. Hepatol. 2021;2021:6696857. doi: 10.1155/2021/6696857. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 82374102/National Natural Science Foundation of China
- 21977021/National Natural Science Foundation of China
- CMEMR2023-B09/State Key Laboratory for Chemistry and Molecular Engineering of Medicinal Resources (Guang-xi Normal University)
- 22201050/National Natural Science Foundation of China
- LZ21H030001/Zhejiang Natural Science Foundation
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

