Molecular Profiling of Peanut under Raw, Roasting, and Autoclaving Conditions Using High-Resolution Magic Angle Spinning and Solution 1H NMR Spectroscopy
- PMID: 38202743
- PMCID: PMC10780471
- DOI: 10.3390/molecules29010162
Molecular Profiling of Peanut under Raw, Roasting, and Autoclaving Conditions Using High-Resolution Magic Angle Spinning and Solution 1H NMR Spectroscopy
Abstract
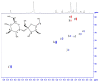
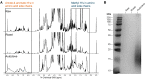
Higher rates of peanut allergy have been observed in countries that commonly roast peanuts prior to consumption. Despite the importance of understanding the role of thermal processing in allergy and on peanut composition, studies toward generating signatures that identify molecular contents following processing are scant. Here, we identified spectral signatures to track changes and differences in the molecular composition of peanuts under raw, roasted, and high-pressure and high-temperature autoclaved conditions. We analyzed both the solid flesh of the seed and solutions derived from soaking peanuts using High-Resolution Magic Angle Spinning (HR-MAS) and solution 1H Nuclear Magnetic Resonance (NMR) spectroscopy, respectively. The NMR spectra of intact peanuts revealed triglycerides as the dominant species, assigned on the basis of multiplets at 4.1 and 4.3 ppm, and corresponding defatted flours revealed the presence of sugars. Sucrose assigned based on a doublet at 5.4 ppm (anomeric proton), and triglycerides were the most abundant small molecules observed, with little variation between conditions. Soaked peanut solutions were devoid of lipids, and their resulting spectra matched the profiles of defatted peanuts. Spectral signatures resulting from autoclaving differed strikingly between those from raw and roasted peanuts, with considerable line-broadening in regions corresponding to proteins and amino-acid side chains, from 0.5 to 2.0 ppm and 6.5 to 8.5 ppm. Taken together, by using complementary NMR methods to obtain a fingerprint of the molecular components in peanuts, we demonstrated that autoclaving led to a distinct composition, likely resulting from the hydrolytic cleavage of proteins, the most important molecule of the allergic reaction.
Keywords: autoclaving; food processing; molecular profiling; nuclear magnetic resonance; peanut.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Influence of thermal processing on the allergenicity of peanut proteins.J Agric Food Chem. 2005 Jun 1;53(11):4547-53. doi: 10.1021/jf050091p. J Agric Food Chem. 2005. PMID: 15913323
-
Different thermal processing effects on peanut allergenicity.J Sci Food Agric. 2019 Mar 30;99(5):2321-2328. doi: 10.1002/jsfa.9430. Epub 2018 Dec 3. J Sci Food Agric. 2019. PMID: 30407639
-
Quantitative Proteomic Profiling of Peanut Allergens in Food Ingredients Used for Oral Food Challenges.Anal Chem. 2016 Jun 7;88(11):5689-95. doi: 10.1021/acs.analchem.5b04466. Epub 2016 May 13. Anal Chem. 2016. PMID: 27064171
-
Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy.Acc Chem Res. 2017 Apr 18;50(4):1105-1113. doi: 10.1021/acs.accounts.7b00082. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28353338 Free PMC article. Review.
-
Peanut (Arachis hypogaea) allergen powder-dnfp for the mitigation of allergic reactions to peanuts in children and adolescents.Expert Rev Clin Immunol. 2023 Mar;19(3):253-265. doi: 10.1080/1744666X.2023.2159812. Epub 2022 Dec 22. Expert Rev Clin Immunol. 2023. PMID: 36524617 Review.
References
-
- Bonku R., Yu J. Health aspects of peanuts as an outcome of its chemical composition. Food Sci. Hum. Wellness. 2020;9:21–30. doi: 10.1016/j.fshw.2019.12.005. - DOI
-
- Han Y., Lin J., Bardina L., Grishina G.A., Lee C., Seo W.H., Sampson H.A. What Characteristics Confer Proteins the Ability to Induce Allergic Responses? IgE Epitope Mapping and Comparison of the Structure of Soybean 2S Albumins and Ara h 2. Molecules. 2016;21:622. doi: 10.3390/molecules21050622. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

