MolOptimizer: A Molecular Optimization Toolkit for Fragment-Based Drug Design
- PMID: 38202859
- PMCID: PMC10780997
- DOI: 10.3390/molecules29010276
MolOptimizer: A Molecular Optimization Toolkit for Fragment-Based Drug Design
Abstract
MolOptimizer is a user-friendly computational toolkit designed to streamline the hit-to-lead optimization process in drug discovery. MolOptimizer extracts features and trains machine learning models using a user-provided, labeled, and small-molecule dataset to accurately predict the binding values of new small molecules that share similar scaffolds with the target in focus. Hosted on the Azure web-based server, MolOptimizer emerges as a vital resource, accelerating the discovery and development of novel drug candidates with improved binding properties.
Keywords: cheminformatics; fragment screening; hit-to-lead optimization.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
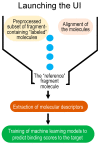

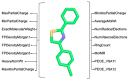
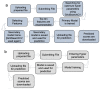

Similar articles
-
From computer-aided drug discovery to computer-driven drug discovery.Drug Discov Today Technol. 2021 Dec;39:111-117. doi: 10.1016/j.ddtec.2021.08.001. Epub 2021 Aug 30. Drug Discov Today Technol. 2021. PMID: 34906321 Review.
-
DeepScreening: a deep learning-based screening web server for accelerating drug discovery.Database (Oxford). 2019 Jan 1;2019:baz104. doi: 10.1093/database/baz104. Database (Oxford). 2019. PMID: 31608949 Free PMC article.
-
Machine Learning guided early drug discovery of small molecules.Drug Discov Today. 2022 Aug;27(8):2209-2215. doi: 10.1016/j.drudis.2022.03.017. Epub 2022 Mar 29. Drug Discov Today. 2022. PMID: 35364270 Review.
-
ACFIS: a web server for fragment-based drug discovery.Nucleic Acids Res. 2016 Jul 8;44(W1):W550-6. doi: 10.1093/nar/gkw393. Epub 2016 May 5. Nucleic Acids Res. 2016. PMID: 27150808 Free PMC article.
-
Sanjeevini: a freely accessible web-server for target directed lead molecule discovery.BMC Bioinformatics. 2012;13 Suppl 17(Suppl 17):S7. doi: 10.1186/1471-2105-13-S17-S7. Epub 2012 Dec 13. BMC Bioinformatics. 2012. PMID: 23282245 Free PMC article.
Cited by
-
Exploring the Promise and Challenges of Artificial Intelligence in Biomedical Research and Clinical Practice.J Cardiovasc Pharmacol. 2024 May 1;83(5):403-409. doi: 10.1097/FJC.0000000000001546. J Cardiovasc Pharmacol. 2024. PMID: 38323891 Free PMC article. Review.
References
-
- Tam B., Sherf D., Cohen S., Eisdorfer S.A., Perez M., Soffer A., Vilenchik D., Akabayov S.R., Wagner G., Akabayov B. Discovery of small-molecule inhibitors targeting the ribosomal peptidyl transferase center (PTC) of M. tuberculosis. Chem. Sci. 2019;10:8764–8767. doi: 10.1039/C9SC02520K. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

