mid-DeepLabv3+: A Novel Approach for Image Semantic Segmentation Applied to African Food Dietary Assessments
- PMID: 38203070
- PMCID: PMC10781344
- DOI: 10.3390/s24010209
mid-DeepLabv3+: A Novel Approach for Image Semantic Segmentation Applied to African Food Dietary Assessments
Abstract
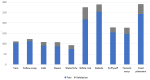
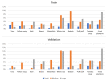
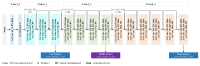
Recent decades have witnessed the development of vision-based dietary assessment (VBDA) systems. These systems generally consist of three main stages: food image analysis, portion estimation, and nutrient derivation. The effectiveness of the initial step is highly dependent on the use of accurate segmentation and image recognition models and the availability of high-quality training datasets. Food image segmentation still faces various challenges, and most existing research focuses mainly on Asian and Western food images. For this reason, this study is based on food images from sub-Saharan Africa, which pose their own problems, such as inter-class similarity and dishes with mixed-class food. This work focuses on the first stage of VBDAs, where we introduce two notable contributions. Firstly, we propose mid-DeepLabv3+, an enhanced food image segmentation model based on DeepLabv3+ with a ResNet50 backbone. Our approach involves adding a middle layer in the decoder path and SimAM after each extracted backbone feature layer. Secondly, we present CamerFood10, the first food image dataset specifically designed for sub-Saharan African food segmentation. It includes 10 classes of the most consumed food items in Cameroon. On our dataset, mid-DeepLabv3+ outperforms benchmark convolutional neural network models for semantic image segmentation, with an mIoU (mean Intersection over Union) of 65.20%, representing a +10.74% improvement over DeepLabv3+ with the same backbone.
Keywords: CNN; CamerFood10 dataset; food segmentation; semantic segmentation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Semantic segmentation of UAV remote sensing images based on edge feature fusing and multi-level upsampling integrated with Deeplabv3.PLoS One. 2023 Jan 20;18(1):e0279097. doi: 10.1371/journal.pone.0279097. eCollection 2023. PLoS One. 2023. PMID: 36662763 Free PMC article.
-
Cattle Target Segmentation Method in Multi-Scenes Using Improved DeepLabV3+ Method.Animals (Basel). 2023 Aug 4;13(15):2521. doi: 10.3390/ani13152521. Animals (Basel). 2023. PMID: 37570328 Free PMC article.
-
A comparative study of pre-trained convolutional neural networks for semantic segmentation of breast tumors in ultrasound.Comput Biol Med. 2020 Nov;126:104036. doi: 10.1016/j.compbiomed.2020.104036. Epub 2020 Oct 8. Comput Biol Med. 2020. PMID: 33059238
-
[Application of semantic segmentation based on convolutional neural network in medical images].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2020 Jun 25;37(3):533-540. doi: 10.7507/1001-5515.201906067. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2020. PMID: 32597097 Free PMC article. Review. Chinese.
-
Applying Image-Based Food-Recognition Systems on Dietary Assessment: A Systematic Review.Adv Nutr. 2022 Dec 22;13(6):2590-2619. doi: 10.1093/advances/nmac078. Adv Nutr. 2022. PMID: 35803496 Free PMC article.
Cited by
-
Lightweight DeepLabv3+ for Semantic Food Segmentation.Foods. 2025 Apr 9;14(8):1306. doi: 10.3390/foods14081306. Foods. 2025. PMID: 40282708 Free PMC article.
References
-
- World Health Organization . Noncommunicable Diseases: Progress Monitor 2022. World Health Organization; Genova, Switzerland: 2022.
-
- Min W., Jiang S., Liu L., Rui Y., Jain R. A survey on food computing. ACM Comput. Surv. 2019;52:1–36. doi: 10.1145/3329168. - DOI
-
- Wang W., Min W., Li T., Dong X., Li H., Jiang S. A review on vision-based analysis for automatic dietary assessment. Trends Food Sci. Technol. 2022;122:223–237. doi: 10.1016/j.tifs.2022.02.017. - DOI
-
- Subhi M.A., Ali S.H., Mohammed M.A. Vision-based approaches for automatic food recognition and dietary assessment: A survey. IEEE Access. 2019;7:35370–35381. doi: 10.1109/ACCESS.2019.2904519. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

