High-Throughput Measure of Mitochondrial Superoxide Levels as a Marker of Coronary Artery Disease to Accelerate Drug Translation in Patient-Derived Endothelial Cells Using Opera Phenix® Technology
- PMID: 38203193
- PMCID: PMC10779289
- DOI: 10.3390/ijms25010022
High-Throughput Measure of Mitochondrial Superoxide Levels as a Marker of Coronary Artery Disease to Accelerate Drug Translation in Patient-Derived Endothelial Cells Using Opera Phenix® Technology
Abstract
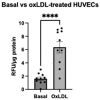
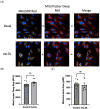
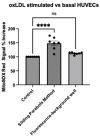
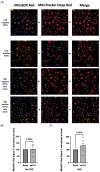
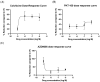
Improved human-relevant preclinical models of coronary artery disease (CAD) are needed to improve translational research and drug discovery. Mitochondrial dysfunction and associated oxidative stress contribute to endothelial dysfunction and are a significant factor in the development and progression of CAD. Endothelial colony-forming cells (ECFCs) can be derived from peripheral blood mononuclear cells (PBMCs) and offer a unique potentially personalised means for investigating new potential therapies targeting important components of vascular function. We describe the application of the high-throughput and confocal Opera Phenix® High-Content Screening System to examine mitochondrial superoxide (mROS) levels, mitochondrial membrane potential, and mitochondrial area in both established cell lines and patient-derived ECFCs simultaneously. Unlike traditional plate readers, the Opera Phenix® is an imaging system that integrates automated confocal microscopy, precise fluorescent detection, and multi-parameter algorithms to visualize and precisely quantify targeted biological processes at a cellular level. In this study, we measured mROS production in human umbilical vein endothelial cells (HUVECs) and patient-derived ECFCs using the mROS production probe, MitoSOXTM Red. HUVECs exposed to oxidized low-density lipoprotein (oxLDL) increased mROS levels by 47.7% (p < 0.0001). A pooled group of patient-derived ECFCs from participants with CAD (n = 14) exhibited 30.9% higher mROS levels compared to patients with no CAD when stimulated with oxLDL (n = 14; p < 0.05). When tested against a small group of candidate compounds, this signal was attenuated by PKT-100 (36.22% reduction, p = 0.03), a novel P2X7 receptor antagonist. This suggests the P2X7 receptor as a valid target against excess mROS levels. As such, these findings highlight the potential of the MitoSOX-Opera Phenix technique to be used for drug discovery efforts in CAD.
Keywords: drug screening; endothelial colony-forming cells; endothelial dysfunction; high-content imaging; high-throughput screening; mROS; mitochondria.
Conflict of interest statement
G.A.F. has received grant support from the National Health & Medical Research Council (Australia), Abbott Diagnostic, Sanofi, Janssen Pharmaceuticals, and NSW Health. G.A.F. has received personal fees from CSL and CPC Clinical Research. G.A.F. serves as a Board Member for the Heart Foundation, President of the Australian Cardiovascular Alliance, Founding Director/CMO of Prokardia, and CSO of CAD Frontiers. G.A.F. has a patent “Patent Biomarkers and Oxidative Stress” which was awarded in the USA in May 2017 (US9638699B2) and licensed to Northern Sydney Local Health District. M.K. and G.A.F are co-founders of Prokardia Pty Ltd. and named inventors on a provisional patent filed by The University of Sydney (Use of P2X7R antagonists in cardiovascular disease; PCT/AU2018/050905) licensed by start-up Prokardia as well as a provisional method-of-use patent for AZD9056– “Adamantyl P2X7 receptor antagonists and their use in the treatment of cardiovascular diseases” (P0061015AU). W.E.L., M.B., E.G., O.T., S.T.V., K.A.K, M.P.G. and S.M.G. report no conflict of interest.
Figures





References
-
- Cardiovascular Diseases (CVDs) [(accessed on 24 April 2023)]. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases...
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

