Gene Characterization of Nocturnin Paralogues in Goldfish: Full Coding Sequences, Structure, Phylogeny and Tissue Expression
- PMID: 38203224
- PMCID: PMC10779419
- DOI: 10.3390/ijms25010054
Gene Characterization of Nocturnin Paralogues in Goldfish: Full Coding Sequences, Structure, Phylogeny and Tissue Expression
Abstract
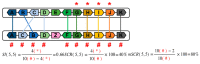
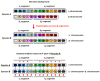
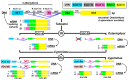
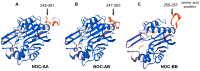
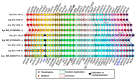
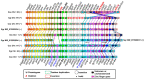
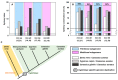
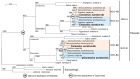
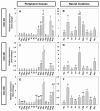
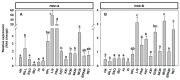
The aim of this work is the full characterization of all the nocturnin (noc) paralogues expressed in a teleost, the goldfish. An in silico analysis of the evolutive origin of noc in Osteichthyes is performed, including the splicing variants and new paralogues appearing after teleostean 3R genomic duplication and the cyprinine 4Rc. After sequencing the full-length mRNA of goldfish, we obtained two isoforms for noc-a (noc-aa and noc-ab) with two splice variants (I and II), and only one for noc-b (noc-bb) with two transcripts (II and III). Using the splicing variant II, the prediction of the secondary and tertiary structures renders a well-conserved 3D distribution of four α-helices and nine β-sheets in the three noc isoforms. A synteny analysis based on the localization of noc genes in the patrilineal or matrilineal subgenomes and a phylogenetic tree of protein sequences were accomplished to stablish a classification and a long-lasting nomenclature of noc in goldfish, and valid to be extrapolated to allotetraploid Cyprininae. Finally, both goldfish and zebrafish showed a broad tissue expression of all the noc paralogues. Moreover, the enriched expression of specific paralogues in some tissues argues in favour of neo- or subfunctionalization.
Keywords: Carassius auratus; gene evolution; molecular cloning; nocturnin nomenclature; phylogenetic tree; protein structure; synteny analysis; synteny quantification; tissue distribution; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
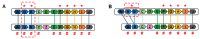




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

