Comparative Analysis of Cyclization Techniques in Stapled Peptides: Structural Insights into Protein-Protein Interactions in a SARS-CoV-2 Spike RBD/hACE2 Model System
- PMID: 38203338
- PMCID: PMC10778704
- DOI: 10.3390/ijms25010166
Comparative Analysis of Cyclization Techniques in Stapled Peptides: Structural Insights into Protein-Protein Interactions in a SARS-CoV-2 Spike RBD/hACE2 Model System
Abstract
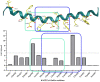
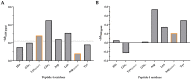
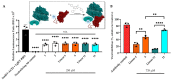
Medicinal chemistry is constantly searching for new approaches to develop more effective and targeted therapeutic molecules. The design of peptidomimetics is a promising emerging strategy that is aimed at developing peptides that mimic or modulate the biological activity of proteins. Among these, stapled peptides stand out for their unique ability to stabilize highly frequent helical motifs, but they have failed to be systematically reported. Here, we exploit chemically diverse helix-inducing i, i + 4 constraints-lactam, hydrocarbon, triazole, double triazole and thioether-on two distinct short sequences derived from the N-terminal peptidase domain of hACE2 upon structural characterization and in silico alanine scan. Our overall objective was to provide a sequence-independent comparison of α-helix-inducing staples using circular dichroism (CD) and nuclear magnetic resonance (NMR) spectroscopy. We identified a 9-mer lactam stapled peptide derived from the hACE2 sequence (His34-Gln42) capable of reaching its maximal helicity of 55% with antiviral activity in bioreporter- and pseudovirus-based inhibition assays. To the best of our knowledge, this study is the first comprehensive investigation comparing several cyclization methods with the goal of generating stapled peptides and correlating their secondary structures with PPI inhibitions using a highly topical model system (i.e., the interaction of SARS-CoV-2 Spike RBD with hACE2).
Keywords: SARS-CoV-2; circular dichroism (CD); nuclear magnetic resonance (NMR); peptidomimetics; protein–protein interaction (PPI).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



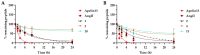
Similar articles
-
Identification of a short ACE2-derived stapled peptide targeting the SARS-CoV-2 spike protein.Eur J Med Chem. 2023 Mar 5;249:115118. doi: 10.1016/j.ejmech.2023.115118. Epub 2023 Jan 17. Eur J Med Chem. 2023. PMID: 36682293 Free PMC article.
-
Targeting SARS-CoV-2 spike protein by stapled hACE2 peptides.Chem Commun (Camb). 2021 Apr 4;57(26):3283-3286. doi: 10.1039/d0cc08387a. Epub 2021 Mar 2. Chem Commun (Camb). 2021. PMID: 33651072
-
Potential chimeric peptides to block the SARS-CoV-2 spike receptor-binding domain.F1000Res. 2020 Jun 9;9:576. doi: 10.12688/f1000research.24074.1. eCollection 2020. F1000Res. 2020. PMID: 32802318 Free PMC article.
-
Stapled peptides as potential inhibitors of SARS-CoV-2 binding to the hACE2 receptor.J Pept Sci. 2022 Sep;28(9):e3409. doi: 10.1002/psc.3409. Epub 2022 Feb 26. J Pept Sci. 2022. PMID: 35165970 Free PMC article. Review.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
Cited by
-
Rerouting therapeutic peptides and unlocking their potential against SARS-CoV2.3 Biotech. 2025 May;15(5):116. doi: 10.1007/s13205-025-04270-0. Epub 2025 Apr 4. 3 Biotech. 2025. PMID: 40191455 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

