Mitochondrial Reactive Oxygen Species in TRIF-Dependent Toll-like Receptor 3 Signaling in Bronchial Epithelial Cells against Viral Infection
- PMID: 38203397
- PMCID: PMC10778811
- DOI: 10.3390/ijms25010226
Mitochondrial Reactive Oxygen Species in TRIF-Dependent Toll-like Receptor 3 Signaling in Bronchial Epithelial Cells against Viral Infection
Abstract
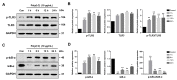
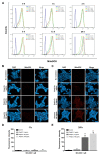
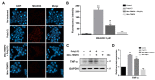
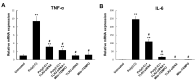
Toll-like receptor 3 (TLR3) plays an important role in double-stranded RNA recognition and triggers the innate immune response by acting as a key receptor against viral infections. Intracellular reactive oxygen species (ROS) are involved in TLR3-induced inflammatory responses during viral infections; however, their relationship with mitochondrial ROS (mtROS) remains largely unknown. In this study, we show that polyinosinic-polycytidylic acid (poly(I:C)), a mimic of viral RNA, induced TLR3-mediated nuclear factor-kappa B (NF-κB) signaling pathway activation and enhanced mtROS generation, leading to inflammatory cytokine production. TLR3-targeted small interfering RNA (siRNA) and Mito-TEMPO inhibited inflammatory cytokine production in poly(I:C)-treated BEAS-2B cells. Poly(I:C) recruited the TLR3 adaptor molecule Toll/IL-1R domain-containing adaptor, inducing IFN (TRIF) and activated NF-κB signaling. Additionally, TLR3-induced mtROS generation suppression and siRNA-mediated TRIF downregulation attenuated mitochondrial antiviral signaling protein (MAVS) degradation. Our findings provide insights into the TLR3-TRIF signaling pathway and MAVS in viral infections, and suggest TLR3-mtROS as a therapeutic target for the treatment of airway inflammatory and viral infectious diseases.
Keywords: TLR3 signaling; bronchial epithelial cells; mitochondria; reactive oxygen species; viral infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

