Harnessing ADAR-Mediated Site-Specific RNA Editing in Immune-Related Disease: Prediction and Therapeutic Implications
- PMID: 38203521
- PMCID: PMC10779106
- DOI: 10.3390/ijms25010351
Harnessing ADAR-Mediated Site-Specific RNA Editing in Immune-Related Disease: Prediction and Therapeutic Implications
Abstract
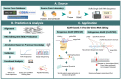
ADAR (Adenosine Deaminases Acting on RNA) proteins are a group of enzymes that play a vital role in RNA editing by converting adenosine to inosine in RNAs. This process is a frequent post-transcriptional event observed in metazoan transcripts. Recent studies indicate widespread dysregulation of ADAR-mediated RNA editing across many immune-related diseases, such as human cancer. We comprehensively review ADARs' function as pattern recognizers and their capability to contribute to mediating immune-related pathways. We also highlight the potential role of site-specific RNA editing in maintaining homeostasis and its relationship to various diseases, such as human cancers. More importantly, we summarize the latest cutting-edge computational approaches and data resources for predicting and analyzing RNA editing sites. Lastly, we cover the recent advancement in site-directed ADAR editing tool development. This review presents an up-to-date overview of ADAR-mediated RNA editing, how site-specific RNA editing could potentially impact disease pathology, and how they could be harnessed for therapeutic applications.
Keywords: ADAR; RNA editing; computational resources; immune-related disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Structural perspectives on adenosine to inosine RNA editing by ADARs.Mol Ther Nucleic Acids. 2024 Jul 19;35(3):102284. doi: 10.1016/j.omtn.2024.102284. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39165563 Free PMC article. Review.
-
Artificial RNA Editing with ADAR for Gene Therapy.Curr Gene Ther. 2020;20(1):44-54. doi: 10.2174/1566523220666200516170137. Curr Gene Ther. 2020. PMID: 32416688 Review.
-
Mechanisms and implications of ADAR-mediated RNA editing in cancer.Cancer Lett. 2017 Dec 28;411:27-34. doi: 10.1016/j.canlet.2017.09.036. Epub 2017 Sep 30. Cancer Lett. 2017. PMID: 28974449 Review.
-
Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity.Nat Methods. 2019 Nov;16(11):1131-1138. doi: 10.1038/s41592-019-0610-9. Epub 2019 Oct 21. Nat Methods. 2019. PMID: 31636457
-
In cancer, A-to-I RNA editing can be the driver, the passenger, or the mechanic.Drug Resist Updat. 2017 May;32:16-22. doi: 10.1016/j.drup.2017.09.001. Epub 2017 Oct 4. Drug Resist Updat. 2017. PMID: 29145975 Review.
Cited by
-
RNA modifications in cancer.MedComm (2020). 2025 Jan 10;6(1):e70042. doi: 10.1002/mco2.70042. eCollection 2025 Jan. MedComm (2020). 2025. PMID: 39802639 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

