Control Theory and Systems Biology: Potential Applications in Neurodegeneration and Search for Therapeutic Targets
- PMID: 38203536
- PMCID: PMC10778851
- DOI: 10.3390/ijms25010365
Control Theory and Systems Biology: Potential Applications in Neurodegeneration and Search for Therapeutic Targets
Abstract
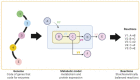
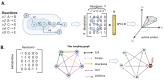
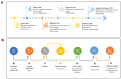
Control theory, a well-established discipline in engineering and mathematics, has found novel applications in systems biology. This interdisciplinary approach leverages the principles of feedback control and regulation to gain insights into the complex dynamics of cellular and molecular networks underlying chronic diseases, including neurodegeneration. By modeling and analyzing these intricate systems, control theory provides a framework to understand the pathophysiology and identify potential therapeutic targets. Therefore, this review examines the most widely used control methods in conjunction with genomic-scale metabolic models in the steady state of the multi-omics type. According to our research, this approach involves integrating experimental data, mathematical modeling, and computational analyses to simulate and control complex biological systems. In this review, we find that the most significant application of this methodology is associated with cancer, leaving a lack of knowledge in neurodegenerative models. However, this methodology, mainly associated with the Minimal Dominant Set (MDS), has provided a starting point for identifying therapeutic targets for drug development and personalized treatment strategies, paving the way for more effective therapies.
Keywords: control theory; genome-scale metabolic networks; neurodegenerative diseases; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Synthesizing Systems Biology Knowledge from Omics Using Genome-Scale Models.Proteomics. 2020 Sep;20(17-18):e1900282. doi: 10.1002/pmic.201900282. Epub 2020 Jul 12. Proteomics. 2020. PMID: 32579720 Free PMC article. Review.
-
Machine learning for the advancement of genome-scale metabolic modeling.Biotechnol Adv. 2024 Sep;74:108400. doi: 10.1016/j.biotechadv.2024.108400. Epub 2024 Jun 27. Biotechnol Adv. 2024. PMID: 38944218 Review.
-
Big data in yeast systems biology.FEMS Yeast Res. 2019 Nov 1;19(7):foz070. doi: 10.1093/femsyr/foz070. FEMS Yeast Res. 2019. PMID: 31603503 Review.
-
Introduction: Cancer Gene Networks.Methods Mol Biol. 2017;1513:1-9. doi: 10.1007/978-1-4939-6539-7_1. Methods Mol Biol. 2017. PMID: 27807826
-
Genome-scale modeling of human metabolism - a systems biology approach.Biotechnol J. 2013 Sep;8(9):985-96. doi: 10.1002/biot.201200275. Epub 2013 Apr 24. Biotechnol J. 2013. PMID: 23613448 Review.
Cited by
-
Redefining Roles: A Paradigm Shift in Tryptophan-Kynurenine Metabolism for Innovative Clinical Applications.Int J Mol Sci. 2024 Nov 27;25(23):12767. doi: 10.3390/ijms252312767. Int J Mol Sci. 2024. PMID: 39684480 Free PMC article. Review.
-
Improvement in the prediction power of an astrocyte genome-scale metabolic model using multi-omic data.Front Syst Biol. 2025 Jan 3;4:1500710. doi: 10.3389/fsysb.2024.1500710. eCollection 2024. Front Syst Biol. 2025. PMID: 40809123 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

