A Plasmid-Borne Gene Cluster Flanked by Two Restriction-Modification Systems Enables an Arctic Strain of Psychrobacter sp. to Decompose SDS
- PMID: 38203722
- PMCID: PMC10779009
- DOI: 10.3390/ijms25010551
A Plasmid-Borne Gene Cluster Flanked by Two Restriction-Modification Systems Enables an Arctic Strain of Psychrobacter sp. to Decompose SDS
Abstract
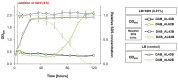
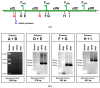
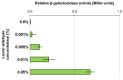
The cold-adapted Psychrobacter sp. strain DAB_AL62B, isolated from ornithogenic deposits on the Arctic island of Spitsbergen, harbors a 34.5 kb plasmid, pP62BP1, which carries a genetic SLF module predicted to enable the host bacterium to metabolize alkyl sulfates including sodium dodecyl sulfate (SDS), a common anionic surfactant. In this work, we experimentally confirmed that the pP62BP1-harboring strain is capable of SDS degradation. The slfCHSL genes were shown to form an operon whose main promoter, PslfC, is negatively regulated by the product of the slfR gene in the absence of potential substrates. We showed that lauryl aldehyde acts as an inducer of the operon. The analysis of the draft genome sequence of the DAB_AL62B strain revealed that the crucial enzyme of the SDS degradation pathway-an alkyl sulfatase-is encoded only within the plasmid. The SLF module is flanked by two restriction-modification systems, which were shown to exhibit the same sequence specificity. We hypothesize that the maintenance of pP62BP1 may be dependent on this unique genetic organization.
Keywords: Psychrobacter; SDS degradation; alkyl sulfatase; psychrophile; restriction–modification system.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





References
-
- Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., Mcveigh R., O’neill K., Robbertse B., et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database J. Biol. Databases Curation. 2020;2020:baaa062. doi: 10.1093/database/baaa062. - DOI - PMC - PubMed
-
- Tratsiak K., Prudnikova T., Drienovska I., Damborsky J., Brynda J., Pachl P., Kuty M., Chaloupkova R., Rezacova P., Smatanova I.K. Crystal structure of the cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5. Pt 5Acta Crystallogr. Sect. F Struct. Biol. Commun. 2019;75:324–331. doi: 10.1107/S2053230X19002796. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

