Exploring the Mechanism of Activation of CFTR by Curcuminoids: An Ensemble Docking Study
- PMID: 38203723
- PMCID: PMC10778693
- DOI: 10.3390/ijms25010552
Exploring the Mechanism of Activation of CFTR by Curcuminoids: An Ensemble Docking Study
Abstract
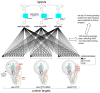
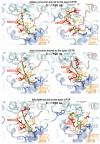
Curcumin, a major constituent of turmeric (Curcuma longa L.), has beneficial effects against several diseases. In cystic fibrosis (CF), this compound improves patients' symptoms by recovering the activity of a number of mutants of the cystic fibrosis transmembrane conductance regulator (CFTR). Despite holding promise in the treatment of CF, the curcumin binding site in CFTR and the molecular mechanism of activation of this channel are still unknown. The results of this study, based on docking and molecular dynamics (MD) simulations, allow us to propose that curcumin binds the closed ATP-free CFTR near the nucleotide-binding domain 1 (NBD1)/ICl1/ICl4 interface. The bound ligand, once approached by the nucleotide-binding domain 2 (NBD2) during transient channel opening, lays at a multiple interdomain cross point. Thereafter, curcumin can bridge NBD1 and NBD2, and also ICL1/ICL4 and ICL2/ICL3, finally tightening the same interdomain interactions that normally uphold the open conformation in the wild-type ATP-bound CFTR. The proposed binding site is compatible with biochemical observations made in previous CFTR-curcumin interaction studies. These findings provide a framework for the design of novel drugs that activate CFTR mutants characterized by defects in ATP binding and/or NBD dimerization or even lacking NBD2.
Keywords: CFTR; CFTR modulators; curcumin; cystic fibrosis; cystic fibrosis transmembrane conductance regulator; docking; molecular dynamics.
Conflict of interest statement
The author declares no conflict of interest.
Figures
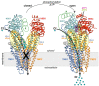






Similar articles
-
Curcumin opens cystic fibrosis transmembrane conductance regulator channels by a novel mechanism that requires neither ATP binding nor dimerization of the nucleotide-binding domains.J Biol Chem. 2007 Feb 16;282(7):4533-4544. doi: 10.1074/jbc.M609942200. Epub 2006 Dec 18. J Biol Chem. 2007. PMID: 17178710
-
Molecular Basis for Fe(III)-Independent Curcumin Potentiation of Cystic Fibrosis Transmembrane Conductance Regulator Activity.Biochemistry. 2015 May 12;54(18):2828-40. doi: 10.1021/acs.biochem.5b00219. Epub 2015 Apr 29. Biochemistry. 2015. PMID: 25867080
-
On the interactions between nucleotide binding domains and membrane spanning domains in cystic fibrosis transmembrane regulator: A molecular dynamic study.Biochimie. 2015 Apr;111:19-29. doi: 10.1016/j.biochi.2015.01.010. Epub 2015 Jan 30. Biochimie. 2015. PMID: 25640670
-
Some like it hot: curcumin and CFTR.Trends Mol Med. 2004 Oct;10(10):473-5. doi: 10.1016/j.molmed.2004.08.001. Trends Mol Med. 2004. PMID: 15464445 Review.
-
Effects of curcumin on ion channels and transporters.Front Physiol. 2014 Mar 11;5:94. doi: 10.3389/fphys.2014.00094. eCollection 2014. Front Physiol. 2014. PMID: 24653706 Free PMC article. Review.
References
-
- Mathews C.J., Tabcharani J.A., Chang X.B., Jensen T.J., Riordan J.R., Hanrahan J.W. Di-basic protein kinase A sites regulate bursting rate and nucleotide sensitivity of the cystic fibrosis transmembrane conductance regulator chloride channel. J. Physiol. 1998;508:365–377. doi: 10.1111/j.1469-7793.1998.365bq.x. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

