A Comprehensive Study of MicroRNA in Baculoviruses
- PMID: 38203774
- PMCID: PMC10778818
- DOI: 10.3390/ijms25010603
A Comprehensive Study of MicroRNA in Baculoviruses
Abstract
Baculoviruses are viral pathogens that infect different species of Lepidoptera, Diptera, and Hymenoptera, with a global distribution. Due to their biological characteristics and the biotechnological applications derived from these entities, the Baculoviridae family is an important subject of study and manipulation in the natural sciences. With the advent of RNA interference mechanisms, the presence of baculoviral genes that do not code for proteins but instead generate transcripts similar to microRNAs (miRNAs) has been described. These miRNAs are functionally associated with the regulation of gene expression, both in viral and host sequences. This article provides a comprehensive review of miRNA biogenesis, function, and characterization in general, with a specific focus on those identified in baculoviruses. Furthermore, it delves into the specific roles of baculoviral miRNAs in regulating viral and host genes and presents structural and thermodynamic stability studies that are useful for detecting shared characteristics with predictive utility. This review aims to expand our understanding of the baculoviral miRNAome, contributing to improvements in the production of baculovirus-based biopesticides, management of resistance phenomena in pests, enhancement of recombinant protein production systems, and development of diverse and improved BacMam vectors to meet biomedical demands.
Keywords: RNA folding; baculovirus; gene regulation; microRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

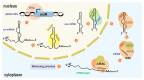



Similar articles
-
Role of microRNAs in insect-baculovirus interactions.Insect Biochem Mol Biol. 2020 Dec;127:103459. doi: 10.1016/j.ibmb.2020.103459. Epub 2020 Sep 19. Insect Biochem Mol Biol. 2020. PMID: 32961323 Review.
-
Protein-Gene Orthology in Baculoviridae: An Exhaustive Analysis to Redefine the Ancestrally Common Coding Sequences.Viruses. 2023 Apr 29;15(5):1091. doi: 10.3390/v15051091. Viruses. 2023. PMID: 37243176 Free PMC article.
-
Baculovirus-Encoded MicroRNAs: A Brief Overview and Future Prospects.Curr Microbiol. 2019 Jun;76(6):738-743. doi: 10.1007/s00284-018-1443-y. Epub 2018 Feb 27. Curr Microbiol. 2019. PMID: 29487989 Review.
-
Solutions against emerging infectious and noninfectious human diseases through the application of baculovirus technologies.Appl Microbiol Biotechnol. 2021 Nov;105(21-22):8195-8226. doi: 10.1007/s00253-021-11615-1. Epub 2021 Oct 7. Appl Microbiol Biotechnol. 2021. PMID: 34618205 Free PMC article. Review.
-
Development of a Dual-Vector System Utilizing MicroRNA Mimics of the Autographa californica miR-1 for an Inducible Knockdown in Insect Cells.Int J Mol Sci. 2019 Jan 27;20(3):533. doi: 10.3390/ijms20030533. Int J Mol Sci. 2019. PMID: 30691228 Free PMC article.
Cited by
-
A myeloid differentiation-like protein in partnership with Toll5 from the pest insect Spodoptera litura senses baculovirus infection.Proc Natl Acad Sci U S A. 2024 Oct 29;121(44):e2415398121. doi: 10.1073/pnas.2415398121. Epub 2024 Oct 23. Proc Natl Acad Sci U S A. 2024. PMID: 39441638 Free PMC article.
-
Characterization of residual microRNAs in AAV vector batches produced in HEK293 mammalian cells and Sf9 insect cells.Mol Ther Methods Clin Dev. 2024 Jul 25;32(3):101305. doi: 10.1016/j.omtm.2024.101305. eCollection 2024 Sep 12. Mol Ther Methods Clin Dev. 2024. PMID: 39220637 Free PMC article.
-
Fungal Pathogen Infection by Metarhizium anisopliae Alters Climbing Behavior of Lymantria dispar with Tree-Top Disease Induced by LdMNPV.Biology (Basel). 2025 Aug 11;14(8):1029. doi: 10.3390/biology14081029. Biology (Basel). 2025. PMID: 40906217 Free PMC article.
References
-
- Rohrmann G.F. Baculovirus Molecular Biology. 4th ed. National Center for Biotechnology Information (US); Bethesda, MD, USA: 2019. - PubMed
-
- Targovnik A.M., Simonin J.A., Mc Callum G.J., Smith I., Cuccovia Warlet F.U., Nugnes M.V., Miranda M.V., Belaich M.N. Solutions against Emerging Infectious and Noninfectious Human Diseases through the Application of Baculovirus Technologies. Appl. Microbiol. Biotechnol. 2021;105:8195–8226. doi: 10.1007/s00253-021-11615-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- PICT-2018 N° 04321/Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación
- PICT-2021-GRFTI-00764/Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación
- PIBAA-2872021010 1251CO/Consejo Nacional de Investigaciones Científicas y Técnicas
LinkOut - more resources
Full Text Sources

