Genome-Wide Identification and Expression Analysis of Chitinase Genes in Watermelon under Abiotic Stimuli and Fusarium oxysporum Infection
- PMID: 38203810
- PMCID: PMC10779513
- DOI: 10.3390/ijms25010638
Genome-Wide Identification and Expression Analysis of Chitinase Genes in Watermelon under Abiotic Stimuli and Fusarium oxysporum Infection
Abstract
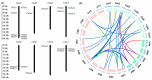
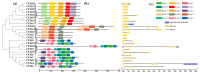
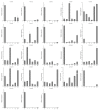
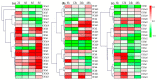
Chitinases, which catalyze the hydrolysis of chitin, the primary components of fungal cell walls, play key roles in defense responses, symbiotic associations, plant growth, and stress tolerance. In this study, 23 chitinase genes were identified in watermelon (Citrullus lanatus [Thunb.]) and classified into five classes through homology search and phylogenetic analysis. The genes with similar exon-intron structures and conserved domains were clustered into the same class. The putative cis-elements involved in the responses to phytohormone, stress, and plant development were identified in their promoter regions. A tissue-specific expression analysis showed that the ClChi genes were primarily expressed in the roots (52.17%), leaves (26.09%), and flowers (34.78%). Moreover, qRT-PCR results indicate that ClChis play multifaceted roles in the interaction between plant/environment. More ClChi members were induced by Race 2 of Fusarium oxysporum f. sp. niveum, and eight genes were expressed at higher levels on the seventh day after inoculation with Races 1 and 2, suggesting that these genes play a key role in the resistance of watermelon to Fusarium wilt. Collectively, these results improve knowledge of the chitinase gene family in watermelon species and help to elucidate the roles played by chitinases in the responses of watermelon to various stresses.
Keywords: Fusarium oxysporum; abiotic stresses; chitinase; expression analyses; genome-wide identification; watermelon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Systematic Genome-Wide Study and Expression Analysis of SWEET Gene Family: Sugar Transporter Family Contributes to Biotic and Abiotic Stimuli in Watermelon.Int J Mol Sci. 2021 Aug 5;22(16):8407. doi: 10.3390/ijms22168407. Int J Mol Sci. 2021. PMID: 34445115 Free PMC article.
-
Genome-wide characterization and expression analysis of the chitinase gene family in chickpea (Cicer arietinum L.) for fungal stress resistance.Mol Biol Rep. 2025 Sep 5;52(1):871. doi: 10.1007/s11033-025-10937-x. Mol Biol Rep. 2025. PMID: 40911260
-
First Report of Fusarium oxysporum f. sp. niveum Race 2 as Causal Agent of Fusarium Wilt of Watermelon in Indiana.Plant Dis. 2005 Jan;89(1):108. doi: 10.1094/PD-89-0108A. Plant Dis. 2005. PMID: 30795300
-
Fusarium oxysporum f. sp. niveum Molecular Diagnostics Past, Present and Future.Int J Mol Sci. 2021 Sep 8;22(18):9735. doi: 10.3390/ijms22189735. Int J Mol Sci. 2021. PMID: 34575897 Free PMC article. Review.
-
Watermelon wilt disease: causes, harms, and control measures.Front Microbiol. 2025 May 7;16:1601130. doi: 10.3389/fmicb.2025.1601130. eCollection 2025. Front Microbiol. 2025. PMID: 40400683 Free PMC article. Review.
Cited by
-
Tandem mass tag (TMT)-based quantitative proteomics analysis reveals the different responses of contrasting alfalfa varieties to drought stress.BMC Genomics. 2024 Aug 27;25(1):806. doi: 10.1186/s12864-024-10702-7. BMC Genomics. 2024. PMID: 39192174 Free PMC article.
-
Genome-Wide Identification and Analysis of Carbohydrate-Binding Modules in Colletotrichum graminicola.Int J Mol Sci. 2025 Jan 22;26(3):919. doi: 10.3390/ijms26030919. Int J Mol Sci. 2025. PMID: 39940689 Free PMC article.
-
Systematic analysis and functional characterization of the chitinase gene family in Fagopyrum tataricum under salt stress.BMC Plant Biol. 2024 Dec 20;24(1):1222. doi: 10.1186/s12870-024-05971-z. BMC Plant Biol. 2024. PMID: 39707214 Free PMC article.
-
Genome-wide identification and biochemical characterization of glycoside hydrolase gene family members in Tilletia Horrida.Mol Biol Rep. 2024 Nov 9;51(1):1136. doi: 10.1007/s11033-024-10059-w. Mol Biol Rep. 2024. PMID: 39520598
-
Bio-priming of tomato seedlings with bacterial consortium against Fusarium oxysporum: a study on morphological parameters and molecular profiling.Front Microbiol. 2025 Jul 9;16:1606896. doi: 10.3389/fmicb.2025.1606896. eCollection 2025. Front Microbiol. 2025. PMID: 40703238 Free PMC article.
References
-
- Pareek N., Vivekanand V., Singh R.P. Advances in Enzyme Biotechnology. Springer; New Delhi, India: 2013. Chitin Deacetylase: Characteristic Molecular Features and Functional Aspects; pp. 125–136.
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

