Specific Circular RNA Signature of Endothelial Cells: Potential Implications in Vascular Pathophysiology
- PMID: 38203852
- PMCID: PMC10779679
- DOI: 10.3390/ijms25010680
Specific Circular RNA Signature of Endothelial Cells: Potential Implications in Vascular Pathophysiology
Abstract
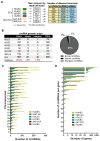
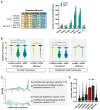
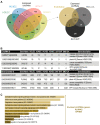
Circular RNAs (circRNAs) are a recently characterized family of gene transcripts forming a covalently closed loop of single-stranded RNA. The extent of their potential for fine-tuning gene expression is still being discovered. Several studies have implicated certain circular RNAs in pathophysiological processes within vascular endothelial cells and cancer cells independently. However, to date, no comparative study of circular RNA expression in different types of endothelial cells has been performed and analysed through the lens of their central role in vascular physiology and pathology. In this work, we analysed publicly available and original RNA sequencing datasets from arterial, veinous, and lymphatic endothelial cells to identify common and distinct circRNA expression profiles. We identified 4713 distinct circRNAs in the compared endothelial cell types, 95% of which originated from exons. Interestingly, the results show that the expression profile of circular RNAs is much more specific to each cell type than linear RNAs, and therefore appears to be more suitable for distinguishing between them. As a result, we have discovered a specific circRNA signature for each given endothelial cell type. Furthermore, we identified a specific endothelial cell circRNA signature that is composed four circRNAs: circCARD6, circPLXNA2, circCASC15 and circEPHB4. These circular RNAs are produced by genes that are related to endothelial cell migration pathways and cancer progression. More detailed studies of their functions could lead to a better understanding of the mechanisms involved in physiological and pathological (lymph)angiogenesis and might open new ways to tackle tumour spread through the vascular system.
Keywords: (lymph)angiogenesis; circRNA; endothelial cell; gene signature.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Merkel Cell Polyomavirus Encodes Circular RNAs (circRNAs) Enabling a Dynamic circRNA/microRNA/mRNA Regulatory Network.mBio. 2020 Dec 15;11(6):e03059-20. doi: 10.1128/mBio.03059-20. mBio. 2020. PMID: 33323517 Free PMC article.
-
Analysis of pig transcriptomes suggests a global regulation mechanism enabling temporary bursts of circular RNAs.RNA Biol. 2019 Sep;16(9):1190-1204. doi: 10.1080/15476286.2019.1621621. Epub 2019 Jun 3. RNA Biol. 2019. PMID: 31120323 Free PMC article.
-
CIRCexplorer pipelines for circRNA annotation and quantification from non-polyadenylated RNA-seq datasets.Methods. 2021 Dec;196:3-10. doi: 10.1016/j.ymeth.2021.02.008. Epub 2021 Feb 12. Methods. 2021. PMID: 33588028
-
CircRNA: a rising star in plant biology.J Genet Genomics. 2022 Dec;49(12):1081-1092. doi: 10.1016/j.jgg.2022.05.004. Epub 2022 May 27. J Genet Genomics. 2022. PMID: 35644325 Review.
-
Biogenesis and Regulatory Roles of Circular RNAs.Annu Rev Cell Dev Biol. 2022 Oct 6;38:263-289. doi: 10.1146/annurev-cellbio-120420-125117. Epub 2022 May 24. Annu Rev Cell Dev Biol. 2022. PMID: 35609906 Free PMC article. Review.
Cited by
-
A Synopsis of Biomarkers in Glioblastoma: Past and Present.Curr Issues Mol Biol. 2024 Jul 3;46(7):6903-6939. doi: 10.3390/cimb46070412. Curr Issues Mol Biol. 2024. PMID: 39057054 Free PMC article. Review.
References
-
- Humphrey J.D., McCulloch A.D. The Cardiovascular System—Anatomy, Physiology and Cell Biology. In: Holzapfel G.A., Ogden R.W., editors. Biomechanics of Soft Tissue in Cardiovascular Systems. Springer; Vienna, Austria: 2003. pp. 1–14. International Centre for Mechanical Sciences.
-
- Pittman R.N. The Circulatory System and Oxygen Transport. Morgan & Claypool Life Sciences; San Rafael, CA, USA: 2011.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

