De Novo Generation and Identification of Novel Compounds with Drug Efficacy Based on Machine Learning
- PMID: 38204214
- PMCID: PMC10962488
- DOI: 10.1002/advs.202307245
De Novo Generation and Identification of Novel Compounds with Drug Efficacy Based on Machine Learning
Abstract
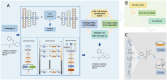
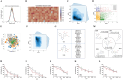
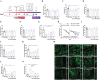
One of the main challenges in small molecule drug discovery is finding novel chemical compounds with desirable activity. Traditional drug development typically begins with target selection, but the correlation between targets and disease remains to be further investigated, and drugs designed based on targets may not always have the desired drug efficacy. The emergence of machine learning provides a powerful tool to overcome the challenge. Herein, a machine learning-based strategy is developed for de novo generation of novel compounds with drug efficacy termed DTLS (Deep Transfer Learning-based Strategy) by using dataset of disease-direct-related activity as input. DTLS is applied in two kinds of disease: colorectal cancer (CRC) and Alzheimer's disease (AD). In each case, novel compound is discovered and identified in in vitro and in vivo disease models. Their mechanism of actionis further explored. The experimental results reveal that DTLS can not only realize the generation and identification of novel compounds with drug efficacy but also has the advantage of identifying compounds by focusing on protein targets to facilitate the mechanism study. This work highlights the significant impact of machine learning on the design of novel compounds with drug efficacy, which provides a powerful new approach to drug discovery.
Keywords: de novo design; drug efficacy; lead compound; machine learning.
© 2024 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Exploring the artificial intelligence and machine learning models in the context of drug design difficulties and future potential for the pharmaceutical sectors.Methods. 2023 Nov;219:82-94. doi: 10.1016/j.ymeth.2023.09.010. Epub 2023 Sep 29. Methods. 2023. PMID: 37778659 Review.
-
Empowering the discovery of novel target-disease associations via machine learning approaches in the open targets platform.BMC Bioinformatics. 2022 Jun 16;23(1):232. doi: 10.1186/s12859-022-04753-4. BMC Bioinformatics. 2022. PMID: 35710324 Free PMC article.
-
De novo generation of dual-target ligands using adversarial training and reinforcement learning.Brief Bioinform. 2021 Nov 5;22(6):bbab333. doi: 10.1093/bib/bbab333. Brief Bioinform. 2021. PMID: 34410338
-
Machine learning prediction of oncology drug targets based on protein and network properties.BMC Bioinformatics. 2020 Mar 14;21(1):104. doi: 10.1186/s12859-020-3442-9. BMC Bioinformatics. 2020. PMID: 32171238 Free PMC article.
-
From machine learning to deep learning: progress in machine intelligence for rational drug discovery.Drug Discov Today. 2017 Nov;22(11):1680-1685. doi: 10.1016/j.drudis.2017.08.010. Epub 2017 Sep 4. Drug Discov Today. 2017. PMID: 28881183 Review.
Cited by
-
Artificial intelligence in lung cancer: current applications, future perspectives, and challenges.Front Oncol. 2024 Dec 23;14:1486310. doi: 10.3389/fonc.2024.1486310. eCollection 2024. Front Oncol. 2024. PMID: 39763611 Free PMC article. Review.
-
Multi-dimensional data-driven computational drug repurposing strategy for screening novel neuroprotective agents in ischemic stroke.Theranostics. 2025 Jun 23;15(15):7653-7676. doi: 10.7150/thno.112608. eCollection 2025. Theranostics. 2025. PMID: 40756350 Free PMC article.
-
From Small Data Modeling to Large Language Model Screening: A Dual-Strategy Framework for Materials Intelligent Design.Adv Sci (Weinh). 2024 Dec;11(45):e2403548. doi: 10.1002/advs.202403548. Epub 2024 Oct 4. Adv Sci (Weinh). 2024. PMID: 39364764 Free PMC article.
-
A Small-Molecule Drug for the Self-Checking of Mitophagy.Angew Chem Int Ed Engl. 2025 Mar 3;64(10):e202421269. doi: 10.1002/anie.202421269. Epub 2025 Jan 22. Angew Chem Int Ed Engl. 2025. PMID: 39800659 Free PMC article.
References
-
- Chan H. C. S., Shan H., Dahoun T., Vogel H., Yuan S., Trends Pharmacol. Sci. 2019, 40, 592. - PubMed
-
- Yang X., Wang Y., Byrne R., Schneider G., Yang S., Chem. Rev. 2019, 19, 10520. - PubMed
-
- Bannigan P., Aldeghi M., Bao Z., Häse F., Aspuru‐Guzik A., Allen C., Adv. Drug Delivery Rev. 2021, 175, 113806. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
