Selection of suitable reference genes for qPCR normalization in different developmental stages of Oenanthe javanica
- PMID: 38205019
- PMCID: PMC10777208
- DOI: 10.3389/fpls.2023.1287589
Selection of suitable reference genes for qPCR normalization in different developmental stages of Oenanthe javanica
Abstract
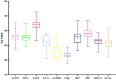
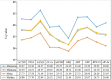
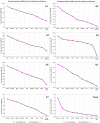
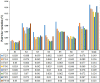
Gene expression analysis is widely used to unravel molecular regulatory mechanisms and identify key genes in plants. Appropriate reference gene is an important prerequisite to ensure the accuracy and reliability of qPCR analysis results. Water dropwort is a plant of the Oenanthe genus in the Apiaceae family, which has high economic benefits. However, the underlying molecular regulatory mechanisms in the growth and development of water dropwort have not been fully understood and the appropriate reference genes in different developmental stages of water dropwort not yet reported. In this study, 10 candidate reference genes (ACTIN, PP2A, SAND, EF-1α, GAPDH, UBQ, MIP, TBP, RPS-18, eIF-4α) were identified and cloned from Oenanthe javanica. The qPCR primers of candidate reference genes were designed and verified. Four statistical algorithms, geNorm, NormFinder, BestKeeper and RefFinder were used to evaluate the expression stability of 10 candidate reference genes in different developmental stages of water dropwort. The results showed that TBP and UBQ were the most stable genes in different developmental stages of water dropwort, while GAPDH was the most unstable gene. The normalization of EXP1 genes at different developmental stages further confirmed the reliability of internal reference genes. The results of this study provide a theoretical basis for selecting appropriate internal reference genes in different developmental stages of water dropwort. This study also provides technical support and reliable basis for the expression analysis of key genes in different developmental stages of water dropwort.
Keywords: RT-qPCR; development stages; growth stages; reference genes; water dropwort.
Copyright © 2023 Feng, Yang, Yan, Sun, Zhou, Liu, Zhao, Wu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Claus L. A., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Prev. Res. (Phila) 64, 5245–5250. doi: 10.1158/0008-5472 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

