Natural flavonoid pectolinarin computationally targeted as a promising drug candidate against SARS-CoV-2
- PMID: 38205118
- PMCID: PMC10776443
- DOI: 10.1016/j.crstbi.2023.100120
Natural flavonoid pectolinarin computationally targeted as a promising drug candidate against SARS-CoV-2
Abstract
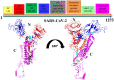
Coronavirus disease-2019 (COVID-19) has become a global pandemic, necessitating the development of new medicines. In this investigation, we identified potential natural flavonoids and compared their inhibitory activity against spike glycoprotein, which is a target of SARS-CoV-2 and SARS-CoV. The target site for the interaction of new inhibitors for the treatment of SARS-CoV-2 has 82% sequence identity and the remaining 18% dissimilarities in RBD S1-subunit, S2-subunit, and 2.5% others. Molecular docking was employed to analyse the various binding processes used by each ligand in a library of 85 natural flavonoids that act as anti-viral medications and FDA authorised treatments for COVID-19. In the binding pocket of the target active site, remdesivir has less binding interaction than pectolinarin, according to the docking analysis. Pectolinarin is a natural flavonoid isolated from Cirsiumsetidensas that has anti-cancer, vasorelaxant, anti-inflammatory, hepatoprotective, anti-diabetic, anti-microbial, and anti-oxidant properties. The S-glycoprotein RBD region (330-583) is inhibited by kaempferol, rhoifolin, and herbacetin, but the S2 subunit (686-1270) is inhibited by pectolinarin, morin, and remdesivir. MD simulation analysis of S-glycoprotein of SARS-CoV-2 with pectolinarin complex at 100ns based on high dock-score. Finally, ADMET analysis was used to validate the proposed compounds with the highest binding energy.
Keywords: Computational analysis; Coronaviruses; Motif; S-glycoproteins; SARS-CoV2.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






Similar articles
-
Potential of Plant Bioactive Compounds as SARS-CoV-2 Main Protease (Mpro) and Spike (S) Glycoprotein Inhibitors: A Molecular Docking Study.Scientifica (Cairo). 2020 Dec 23;2020:6307457. doi: 10.1155/2020/6307457. eCollection 2020. Scientifica (Cairo). 2020. PMID: 33425427 Free PMC article.
-
Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro.J Enzyme Inhib Med Chem. 2020 Dec;35(1):1539-1544. doi: 10.1080/14756366.2020.1801672. J Enzyme Inhib Med Chem. 2020. PMID: 32746637 Free PMC article.
-
Structural Bioinformatics Used to Predict the Protein Targets of Remdesivir and Flavones in SARS-CoV-2 Infection.Med Chem. 2022;18(3):382-393. doi: 10.2174/1573406417666210806154129. Med Chem. 2022. PMID: 34365955
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
-
Identification of Kaempferol as Viral Entry Inhibitor and DL-Arginine as Viral Replication Inhibitor from Selected Plants of Indian Traditional Medicine against COVID-19: An in silico Guided in vitro Approach.Curr Comput Aided Drug Des. 2023;19(4):313-323. doi: 10.2174/1573409919666230112123213. Curr Comput Aided Drug Des. 2023. PMID: 36635906
Cited by
-
Advancements in Antiviral Drug Development: Comprehensive Insights into Design Strategies and Mechanisms Targeting Key Viral Proteins.J Microbiol Biotechnol. 2024 Jul 28;34(7):1376-1384. doi: 10.4014/jmb.2403.03008. Epub 2024 Apr 29. J Microbiol Biotechnol. 2024. PMID: 38934770 Free PMC article. Review.
References
-
- Burak M., Imen Y. Flavonoids and their antioxidant properties. Turkiye Klin Tip Bil Derg. 1999;19(1):296–304.
LinkOut - more resources
Full Text Sources
Miscellaneous

